CMSDAS Pre-Exercise 1: Unix basics
Overview
Teaching: 25 min
Exercises: 5 minQuestions
How do I get started with Unix?
How do I navigate the filesystem?
How do I manipulate files and folder?
Objectives
Setup a working Unix environment on your laptop.
Learn how to use folders and files in Unix.
If you have never used Unix before, please follow the lesson on the HEP Software Foundation website, through at least “3. Working With Files and Directories.” The remainder of the pre-exercises takes place in Unix, so you will need to be familiar with the basics.
Question 1.1
All CMSDAS@LPC 2026 participants should submit answers to the questions in these exercises. Each lesson has it’s own Google form. Head to CMSDAS@LPC2026 Google form 1. The first question verifies that you have a working Unix environment on your personal laptop. Copy-and-paste the following into your shell:
echo "$(whoami) $(date)"This should print your username followed by the current date and time. Copy-and-paste the output into the Google form.
Key Points
Unix is the primary OS used in HEP, CMS included.
CMSDAS Pre-Exercise 2: Using the cmslpc cluster
Overview
Teaching: 0 min
Exercises: 10 minQuestions
Learn how to use the CMSLPC cluster
Objectives
Learn how to use the CMSLPC cluster
Questions
For this lesson, please submit your answers using CMSDAS@LPC2026 Google Form 2.
Introduction
In the previous lesson, you learned how to use Unix on your personal computer. However, the CMS detector produces more than 500 terabytes of data per second, so there’s only so much you can do on your laptop. In this lesson, we will learn how to use the cmslpc computing cluster, which you will use extensively during CMSDAS and possibly for your actual analysis work as well. Information for lxplus users is also provided, where appropriate.
The basics
The cmslpc cluster consists of a large number of “interactive nodes,” which users login to via SSH, and an even larger number of “worker nodes,” which are used for large-scale “batch” computing.
We will use the AlmaLinux 9 operating system (OS), a community-supported OS that is binary-compatible with Red Hat Enterprise Linux (RHEL).
AlmaLinux 8 was chosen for CMS Run 3 data processing, and we are gradually migrating towards the newer AlmaLinux 9. See these slides for the gory details).
CMS users are allocated storage space in a few places:
- (1) a small home directory (2 GB) at
/uscms/home/username, ; - (2) a medium storage directory (200 GB, not backed up!) at
/uscms_data/d[1-3]/username, which is softlinked in your home directory at/uscms/home/username/nobackup, and - (3) a large storage directory on EOS (2 TB) (special filesystem, more info later in this lesson).
The lxplus cluster is configured similarly, with slightly different paths and quotas allocated to users (note that lxplus.cern.ch is an alias for lxplus9.cern.ch, a login node running AlmaLinux 9 OS; use lxplus8.cern.ch to get on a AlmaLinux 8 node.
Logging in
Let’s try logging in to cmslpc using SSH. SSH is a very widely used program for logging into remote Unix clusters; you can check out the HSF SSH exercise to learn more, but for now you can just follow the commands in this exercise. The authentication for cmslpc uses kerberos (your university cluster may allow simple password login or certificate login, which are not covered here).
First, if you have not configured SSH and kerberos on your own computer, you will need to set up a Kerberos configuration file (krb5.conf) and a SSH configuration file (~/.ssh/config). You can follow the prerequisites section here. Once you have the cmslpc-specific SSH and kerberos configuration, execute the following command in the terminal on your own computer:
kinit <YourUsername>@FNAL.GOV
# kinit <YourUsername>@CERN.CH for lxplus users
Enter the kerberos password for your Fermilab account. You now have login access to cmslpc for 24 hours. If you get an error message, double-check that you configure kerberos properly, or head to Mattermost for help.
Next, execute the following to login:
ssh -Y <YourUsername>@cmslpc-el9.fnal.gov
# ssh -Y <YourUsername>@lxplus.cern.ch for lxplus users
If you see a welcome message followed by a command prompt, congratulations, you have successfully logged in! The commands you enter into the command prompt will now be executed on the cmslpc interactive node. If you see an error message instead, something has probably gone wrong with the authentication; please head to Mattermost and post your error message, and an instructor can help you out.
Running simple commands on cmslpc
Note: this exercise will only work on cmslpc-el9.
In this exercise, we will run a few simple commands on cmslpc. At the end, you will type an answer into a spreadsheet (experienced users should feel free to breeze through, but please do upload your answer so we can follow everyone’s progress).
First, make a folder for your CMSDAS work, and cd into that directory:
mkdir ~/nobackup/cmsdas
cd nobackup/cmsdas
Use the following command to copy the runThisCommand.py script:
cp ~cmsdas/preexercises/runThisCommand.py .
Next, copy and paste the following and then hit return:
python3 runThisCommand.py "asdf;klasdjf;kakjsdf;akjf;aksdljf;a" "sldjfqewradsfafaw4efaefawefzdxffasdfw4ffawefawe4fawasdffadsfef"
The response should be your username followed by alphanumeric string of characters unique to your username, for example for a user named gbenelli:
success: gbenelli toraryyv
If you executed the command without copy-pasting (i.e. only ./runThisCommand.py without the additional arguments) the command will return:
Error: You must provide the secret key
Alternatively, copying incorrectly (i.e. different arguments) will return:
Error: You didn't paste the correct input string
If you are not running on cmslpc-el9 (for example locally on a laptop), trying to run the command will result in:
bash: ./runThisCommand.py: No such file or directory
or (for example):
Unknown user: gbenelli.
Question 2.1
Copy-and-paste the alphanumeric string of characters unique to your username in the Google form.
Editing files on cmslpc
The purpose of this exercise is to ensure that the user can edit files. We will first copy and then edit the editThisCommand.py script.
Users of cmslpc have several options for editing remote files. Here are a few examples:
- Edit files directly on cmslpc using a terminal-based code editor, such as
nano,emacs(opens a GUI by default, which is slow over SSH; useemacs -nwto get a terminal-based editor instead), orvim.emacsandvim, in particular, have lots of features for code editing, but also have a steep learning curve. - Edit files on your own computer in the terminal (with the same programs), and upload using, e.g.,
sftp myscript.py username@cmslpc-el9.fnal.gov:my/folder. - Use an application like Visual Studio Code or Sublime Text, either directly on cmslpc (using a remote filesystem plugin, which makes your directory on cmslpc appear as a folder on your computer) or on your own computer (using an SSH or SFTP plugin to automatically upload files to cmslpc). These also have lots of features, and are easier to learn than
emacsorvim.
For the sake of this lesson, will will simply edit a file directly on cmslpc, using nano, emacs, or vim.
If you choose vim, watch this video tutorial or play with this interactive tutorial.
On the cmslpc-el9 cluster, run:
cd ~/nobackup/cmsdas
cp ~cmsdas/preexercises/editThisCommand.py .
Then open editThisCommand.py with your favorite editor (e.g. emacs -nw editThisCommand.py).
The 11th line of this python script throws an error, meaning that python prints an error message and ends the program immediately.
For this exercise, add a # character at the start of the 11th line, which turns the line of code into a comment that is skipped by the python interpreter.
Specifically, change:
# Please comment the line below out by adding a '#' to the front of
# the line.
raise RuntimeError("You need to comment out this line with a #")
to:
# Please comment the line below out by adding a '#' to the front of
# the line.
#raise RuntimeError("You need to comment out this line with a #")
(For vim users: you need to press i to insert text.) Save the file (e.g. in emacs, type ctrl+x ctrl+s to save, ctrl+x ctrl+c to quit the editor; in vim, press ESC to exit insert mode, the “:wq” to save and exit) and execute the command:
./editThisCommand.py
If this is successful, the result will be:
success: gbenelli 0x6D0DB4E0
If the file has not been successfully edited, an error message will result such as:
Traceback (most recent call last):
File "./editThisCommand.py", line 11, in ?
raise RuntimeError, "You need to comment out this line with a #"
RuntimeError: You need to comment out this line with a #
Question 2.2
Copy-and-paste the line beginning with “success”, resulting from the execution of
./editThisCommand.py, into the Google form.
Using the EOS filesystem
Your biggest storage on cmslpc is the EOS filesystem, which behaves differently from a normal Unix folder.
EOS has dedicated commands for interacting with the filesystem, rather than the usual ls, cp, mkdir, etc.
Also, files are addressed using so-called “logical filenames” (LFNs), which you can think of as their location inside EOS, rather than their absolute location (or physical filename, PFN).
The LFNs usually start with /store/....
Click here for the full documentation; here are a few essential commands.
- On cmslpc, the equivalent of
lsiseosls: for example,eosls /store/user/cmsdas/preexercises/. This is actually a cmslpc-specific alias foreos root://cmseos.fnal.gov ls; on other clusters, you’ll have to use this full command. A useful flag iseosls -alh, which will print folder and file sizes. - Similarly to
ls, the cmslpc-specific equivalents ofmkdirandmvareeosmkdirandeosmv. (You can doalias eosmkdirto see the full command behind the alias.) - The equivalent of
cpisxrdcp: for example,xrdcp root://cmseos.fnal.gov//store/user/cmsdas/preexercises/DYJetsToLL_M50_NANOAOD.root .. Theroot://cmseos.fnal.govbit tellsxrdcpwhich EOS instance to use (only one instance for cmslpc users; lxplus has several, e.g.,root://eoscms.cern.chfor CMS data androot://eosuser.cern.chfor user data).
Question 2.3
We will copy a small file from EOS to your nobackup area, containing 10,000 simulated $Z\rightarrow\mu^+\mu^-$ events in the CMS NanoAOD format. We will use this file in later exercises, so make sure not to lose track of it.
Execute the following:
cd ~/nobackup/cmsdas/ xrdcp root://cmseos.fnal.gov//store/user/cmsdas/preexercises/DYJetsToLL_M50_NANOAOD.root .Using
ls -lh DYJetsToLL_M50_NANOAOD.root, how big is this file? (It’s the large number.) Write the answer in the Google form.
Key Points
Learn how to use the CMSLPC cluster
CMSDAS Pre-Exercise 3: ROOT and python basics
Overview
Teaching: 0 min
Exercises: 60 minQuestions
Objectives
Learn how to use ROOT and python
Use ROOT and python to inspect a CMS NanoAOD file
Questions
For this lesson, please submit your answers for the CMSDAS@LPC2026 Google Form 3.
Python and ROOT
Python and ROOT are two of the most important software tools in HEP. If you have never used python before, we highly recommend you go through a tutorial, for example the HSF python lesson. If you are comfortable with basic python, feel free to proceed, as you will learn by doing in the following exercises.
For ROOT, please follow the lesson on the HEP Software Foundation website for ROOT, up through at least the 3rd lesson, 02-tfile-read-write-ttrees.ipynb (of course, you can keep going and learn about RDataFrames, but we won’t use them here). We recommend you click the SWAN button, which opens a session in CERN’s “Service for Web-based ANalysis.” From the service, you can open and run code through Jupyter notebooks, all inside the web browser.
Inspect a NanoAOD file with ROOT
Once you’re comfortable with python and ROOT, let’s go back to cmslpc and look at some real CMS data. Login to the cluster again from your computer:
kinit <YourUsername>@FNAL.GOV
ssh -Y <YourUsername>@cmslpc-el9.fnal.gov
By default, your shell environment will not have ROOT, and will have some very old version of python. A quick way to setup ROOT is to use the LCG (LHC Computing Grid) releases. All you have to do is execute the following script (once per login):
source /cvmfs/sft.cern.ch/lcg/views/LCG_106a/x86_64-el9-gcc14-opt/setup.sh
First, let’s inspect a ROOT file using ROOT’s built-in C++ interpreter, CINT. Run the following to launch ROOT and simultaneously open a file (the same file we copied from EOS in the previous exercise).
cd ~/nobackup/cmsdas
root -l DYJetsToLL_M50_NANOAOD.root
# Note: the -l option stops ROOT from displaying its logo image, which is very slow over SSH
CINT is a quick way to inspect files (to exit CINT/ROOT, just type .q). First, let’s see what’s in the file. Enter _file0->ls() into the interpreter:
root [1] _file0->ls()
TFile** DYJetsToLL_M50_NANOAOD.root
TFile* DYJetsToLL_M50_NANOAOD.root
KEY: TTree Events;1 Events
The file contains just one interesting object, a TTree named Events. Next, let’s check the basic contents of Events. For the number of events in the TTree:
root [4] Events->GetEntries()
(long long) 10000
For the branches in Events, try:
root [6] Events->Print()
******************************************************************************
*Tree :Events : Events *
*Entries : 10000 : Total = 65055196 bytes File Size = 19389904 *
* : : Tree compression factor = 3.33 *
******************************************************************************
*Br 0 :run : run/i *
*Entries : 10000 : Total Size= 40621 bytes File Size = 410 *
*Baskets : 2 : Basket Size= 29696 bytes Compression= 97.91 *
*............................................................................*
*Br 1 :luminosityBlock : luminosityBlock/i *
*Entries : 10000 : Total Size= 40693 bytes File Size = 493 *
*Baskets : 2 : Basket Size= 29696 bytes Compression= 81.48 *
*............................................................................*
*Br 2 :event : event/l *
*Entries : 10000 : Total Size= 80641 bytes File Size = 20690 *
*Baskets : 2 : Basket Size= 58880 bytes Compression= 3.87 *
...
...
...
*............................................................................*
*Br 1626 :HLTriggerFinalPath : HLTriggerFinalPath/O *
*Entries : 10000 : Total Size= 10705 bytes File Size = 280 *
*Baskets : 2 : Basket Size= 8192 bytes Compression= 36.34 *
*............................................................................*
*Br 1627 :L1simulation_step : L1simulation_step/O *
*Entries : 10000 : Total Size= 10699 bytes File Size = 279 *
*Baskets : 2 : Basket Size= 8192 bytes Compression= 36.46 *
*............................................................................*
This prints out a super long list of branches, because NanoAOD assigns a branch for every trigger in CMS. You can filter to looks at, e.g., only muon branches:
root [8] Events->Print("Muon*")
******************************************************************************
*Tree :Events : Events *
*Entries : 10000 : Total = 65055196 bytes File Size = 19389904 *
* : : Tree compression factor = 3.33 *
******************************************************************************
*Br 0 :Muon_dxy : Float_t dxy (with sign) wrt first PV, in cm *
*Entries : 10000 : Total Size= 67214 bytes File Size = 29269 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 2.28 *
*............................................................................*
*Br 1 :Muon_dxyErr : Float_t dxy uncertainty, in cm *
*Entries : 10000 : Total Size= 67222 bytes File Size = 21835 *
*Baskets : 4 : Basket Size= 32000 bytes Compression= 3.05 *
...
Question 3.1
The method
Long64_t TTree:GetEntries (const char *selection)accepts a selection string, and returns the number of events passing the selection criteria (note: written in C++). Use this method to get the number of events with two reconstructed muons:root [0] Events->GetEntries("nMuon >= 2")Write the number of events with at least 2 muon candidates in the Google form.
Plotting with pyROOT
You can also use ROOT in python, almost identically to CINT except with python instead of C++.
(This is possible because of pyROOT, a wrapper around ROOT that creates a nearly 1-to-1 map of all the C++ classes to python classes.)
Make sure you’re logged into cmslpc, and that you have called the LCG setup script in the session.
Then, let’s reopen the NanoAOD file in python. Start a python interactive session by entering python3 (type ctrl-d or exit() to quit)
python3
then enter the following into the python interpreter:
import ROOT
f = ROOT.TFile("DYJetsToLL_M50_NANOAOD.root", "READ")
f.ls()
You should see the same contents of the file as before.
Long64_t TTree::Draw(const char * varexp, const char * selection, ...) is a fast way to make a plot from a TTree.
The first argument, varexp, is what you want to plot (accepts single branches as well as expressions; the string is compiled on the fly by CINT).
The second argument, selection, allows you to filter events before plotting.
Let’s use this function to plot the generator-level Z boson mass distribution, i.e., the truth particles generated by Madgraph (versus the reconstructed particles after the CMS detector simulation has been run).
The branch GenPart_mass contains the mass of all generator-level particles.
The branch GenPart_pdgId contains the so-called PDG ID of the generator-level particles; Z bosons are assigned a PDG ID of 23.
events = f.Get("Events")
c = ROOT.TCanvas()
events.Draw("GenPart_mass", "GenPart_pdgId==23")
c.Draw()
A plot of the Z boson mass should appear, with a mean value close to the Z boson mass of 91.1876 GeV. (If no plot appears, something is probably wrong with the X window system that displays graphical windows over SSH. Following the cmslpc instructions, make sure you have an X windows program installed on your computer, and that you logged in using ssh -Y; ask Mattermost for more help.)
Question 3.2: Z mass plot
The plot includes a “stat box” with basic information about the plotted histogram. Please fill in the mean of the distribution in the Google form.
Key Points
ROOT and python are two key software tools in HEP.
Many CMS analyses use the NanoAOD format, which are simple ROOT ntuples that can be analyzed with standalone ROOT or pyROOT.
There are numerous ways to use ROOT, including the build-in command line interface (based on CINT, a C++ interpreter), pyROOT, Jupyter notebooks, compiled C++, and more.
CMSDAS Pre-Exercise 4: Using git
Overview
Teaching: 0 min
Exercises: 30 minQuestions
How do I setup git on my computer/cluster?
How do I collaborate using GitHub?
Objectives
Setup your git configuration for a given computer.
Learn how to make and commit changes to a git repository.
Learn how to create a pull request on GitHub.
Questions
Please post your answers to the questions in CMSDAS@LPC2026 Google form 4.
Introduction
This exercise is intended to provide you with basic familiarity with Git and GitHub for personal and collaborative use, including terminology, commands, and user interfaces. The exercise proceeds step-by-step through a standard collaboration “Fork and Pull” workflow. This is a highly condensed version of the tutorial exercises at CMSGitTutorial. Students are encouraged to explore those more in-depth exercises if they want to learn more about using Git. There are also accompanying slides on that twiki page. Students with no experience using Git or other version control software are recommended to read at least the first set of slides.
Warning
As a prerequisite for this exercise, please make sure that you have correctly followed the instructions for obtaining a GitHub account in the setup instructions.
Learning Git and GitHub
Git Configuration
If you haven’t setup git on yet, execute the following three commands (note: you should probably do this both on cmslpc and your own laptop, plus any other clusters or computers you use for development).
git config --global user.name "[Name]"
git config --global user.email [Email]
git config --global user.github [Account]
Make sure you replace [Name], [Email], and [Account] with the values corresponding to your GitHub account. After this, you can check the contents of .gitconfig by doing:
cat ~/.gitconfig
Output
[user] name = [Name] email = [Email] github = [Account]
Optional settings:
- Your preferred editor:
git config --global core.editor [your preferred text editor]
- This setting makes Git push the current branch by default, so only the command
git push originis needed. (NOTE: do not try to execute that command now; it will not work without a local repository, which you have not created yet.)
git config --global push.default current
- This is an alias to make the print out of the log more concise and easier to read.
git config --global alias.lol 'log --graph --decorate --pretty=oneline --abbrev-commit'
- These make it easier to clone repositories from GitHub or CERN GitLab, respectively. For example,
git clone github:GitHATSLPC/GitHATS.git.
git config --global url."git@github.com:".insteadOf github:
git config --global url."ssh://git@gitlab.cern.ch:7999/".insteadOf gitlab:
GitHub User Interface
Look carefully at the GitHub user interface on the main page for the GitHATSLPC/GitHATS repository. Click on various tabs.
-
Top left row: Code, Issues, Pull Requests, Actions, Projects, Wiki, Security, Insights

-
Top right row: Notifications, Star, Fork
-
Lower row on Code page: commits, branches, releases, contributors

Collaboration on GitHub
Fork the repository GitHATSLPC/GitHATS repository by clicking “Fork” at the top right corner of the page. This makes a copy of the repository under your GitHub account.
Clone your fork of the repository to a scratch directory on your local machine or cmslpc:
mkdir scratch
cd scratch
git clone git@github.com:[user]/GitHATS.git
Output
Cloning into 'GitHATS'... Enter passphrase for key '/home/------/.ssh/id_rsa': remote: Counting objects: 21, done. remote: Total 21 (delta 0), reused 0 (delta 0), pack-reused 21 Receiving objects: 100% (21/21), done. Resolving deltas: 100% (5/5), done. Checking connectivity... done.
What does the ls command show?
cd GitHATS
ls -a
Output
. .. .git README.md standard_model.md
The .git folder contains a full local copy of the repository.
Inspect the .git directory:
ls .git
Output
config description HEAD hooks index info logs objects packed-refs refs
When you use git clone as we did above, it starts your working area on the default branch for the repository. In this case, that branch is main. (The default branch for a repo can be changed in the “Branches” section of the GitHub settings page, which you explored in the previous step.)
Inspect the branches of the repository.
git branch -a
Output
* main remotes/origin/HEAD -> origin/main remotes/origin/atlas_discovery remotes/origin/cms_discovery remotes/origin/dune_discovery remotes/origin/main
Adding remotes and synchronizing
Look at your remote(s):
git remote
Output
origin
Hint
For additional information you can add the
-voption to the commandgit remote -vOutput
origin git@github.com:[user]/GitHATS.git (fetch) origin git@github.com:[user]/GitHATS.git (push)
The “origin” remote is set by default when you use git clone. Because your repository is a fork, you also want to have a remote that points to the original repo, traditionally called “upstream”.
Add the upstream remote and inspect the result:
git remote add upstream git@github.com:GitHATSLPC/GitHATS.git
git remote -v
Output
origin git@github.com:[user]/GitHATS.git (fetch) origin git@github.com:[user]/GitHATS.git (push) upstream git@github.com:GitHATSLPC/GitHATS.git (fetch) upstream git@github.com:GitHATSLPC/GitHATS.git (push)
Before you make edits to your local repo, you should make sure that your fork is up to date with the main repo. (Someone else might have made some updates in the meantime.)
Check for changes in upstream:
git pull upstream main
Output
From github.com:GitHATSLPC/GitHATS * branch main -> FETCH_HEAD * [new branch] main -> upstream/main Already up-to-date.
Note
git pull upstream mainis equivalent to the following two commands:git fetch upstream main git merge upstream/main
If you pulled any changes from the upstream repository, you should push them back to origin. (Even if you didn’t, you can still practice pushing; nothing will happen.)
Push your local main branch back to your remote fork:
git push origin main
Output
Everything up-to-date
Making edits and committing
When collaborating with other developers on GitHub, it is best to make a separate topic branch to store any changes you want to submit to the main repo. This way, you can keep the default branch in your fork synchronized with upstream, and then make another topic branch when you want to make more changes.
Make a topic branch:
git checkout -b MyBranch
Edit the table standard_model.md to add a new particle. The new particle is called a Giton, with symbol G, spin 2, charge 0, and mass 750 GeV.
Note
Any resemblance to any other real or imaginary particles is entirely coincidental.
Once you have made changes in your working area, you have to stage the changes and then commit them. First, you can inspect the status of your working area.
Try the following commands to show the status:
git status
Output
On branch MyBranch Changes not staged for commit: (use "git add ..." to update what will be committed) (use "git checkout -- ..." to discard changes in working directory) modified: standard_model.md no changes added to commit (use "git add" and/or "git commit -a")
git status -s
Output
M standard_model.md
git diff
Output
diff --git a/standard_model.md b/standard_model.md index 607b7b6..68f37ad 100644 --- a/standard_model.md +++ b/standard_model.md @@ -18,4 +18,5 @@ The Standard Model of Particle Physics | Z boson | Z | 1 | 0 | 91.2 | | W boson | W | 1 | ±1 | 80.4 | | gluon | g | 1 | 0 | 0 | -| Higgs boson | H | 0 | 0 | 125 | \ No newline at end of file +| Higgs boson | H | 0 | 0 | 125 | +| Giton | G | 2 | 0 | 750 |
Now stage your change, and check the status:
git add standard_model.md
git status -s
Output
M standard_model.md
Commit your change:
git commit -m "add Giton to standard model"
Output
[MyBranch b9bc2ce] add Giton to standard model 1 file changed, 2 insertions(+), 1 deletion(-)
Push your topic branch, which now includes the new commit you just made, to origin:
git push origin MyBranch
Output
Enumerating objects: 5, done. Counting objects: 100% (5/5), done. Delta compression using up to 8 threads Compressing objects: 100% (3/3), done. Writing objects: 100% (3/3), 356 bytes | 356.00 KiB/s, done. Total 3 (delta 1), reused 0 (delta 0) remote: Resolving deltas: 100% (1/1), completed with 1 local object. remote: remote: Create a pull request for 'MyBranch' on GitHub by visiting: remote: https://github.com/mtonjes/GitHATS/pull/new/MyBranch remote: To github.com:mtonjes/GitHATS.git * [new branch] MyBranch -> MyBranch
Making a pull request
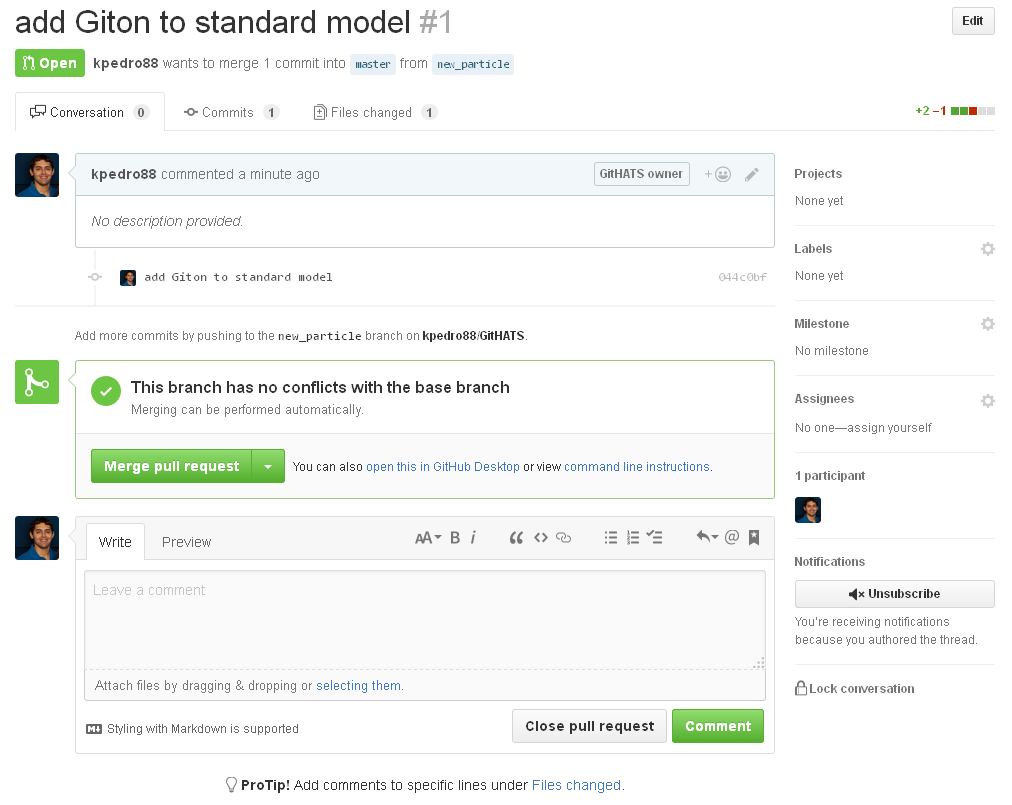
Now that you have made your change, you can submit it for inclusion in the central repository.
When you open the web page (the link listed on the command line above to your own fork) to send a pull request on GitHub, you will notice that you can send a pull request to any fork of the repo (and any branch).
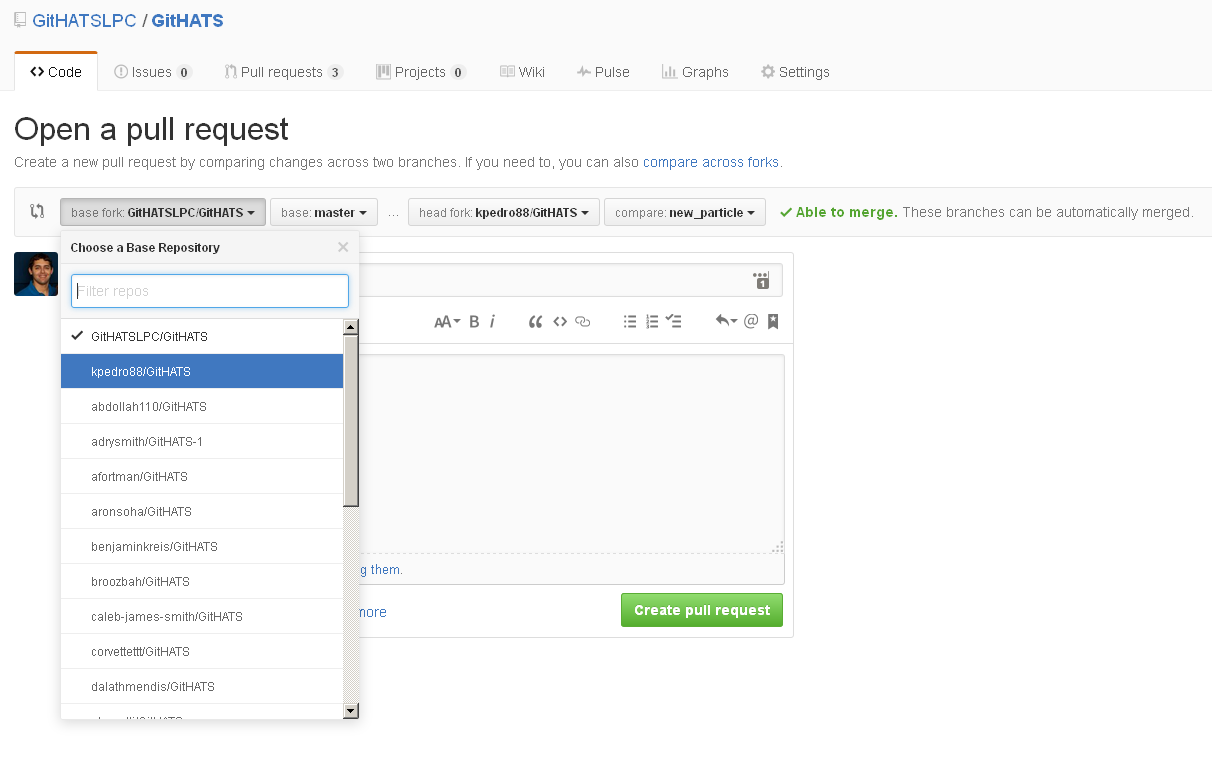
Send a pull request to the main branch of the upstream repo (GitHATSLPC).
Question 4.1
Post the link to your pull request.
For CMSDAS@LPC 2026 please submit your answer at the CMSDAS@LPC2026 Google Form set 4.
Optional
If you want to practice merging a pull request, you can send a pull request from your branch
MyBranchto your own main branch.
Advanced topics
Advanced topics not explored in this exercise include: merging, rebasing, cherry-picking, undoing, removing binary files, and CMSSW-specific commands and usage.
If you want to learn more, here are some additional resources:
Students are encouraged to explore these topics on their own at CMSGitTutorial.
Key Points
Interact with your git configuration using
git config --global.Use the
git clonecommand to obtain a local copy of a git repository.Add and interact with new remotes using the
git remotecommand.Use the
addandcommitcommands to add changes to the local repository.The
pullandpushcommands will transfer changes between the remote and local copies of the repository.
CMSDAS Pre-Exercise 5: CMSSW basics
Overview
Teaching: 0 min
Exercises: 60 minQuestions
Objectives
Setup a CMSSW environment
Use git to download an example EDAnalyzer
Run a CMSSW job on real dimuon data to plot the Z peak
Questions
For this lesson, please submit your answers using CMSDAS@LPC2026 Google Form 5.
CMSSW
CMSSW is CMS’s software framework for analyzing the data produced by the CMS detector. The framework contains a large number of modules (C++), which perform tasks like:
- Loading RAW data (bits from the detector) into nice C++ objects;
- Reconstructing detector hits, physics objects (tracks, electrons, muons, hadrons, jets, …);
- Loading calibrations from databases and applying to physics objects;
- Interfacing with external generator programs like Pythia and Madgraph_aMC@NLO;
- Lots and lots of other things.
Why do you need CMSSW?
With the advent of NanoAOD, a simple ROOT format that does need CMSSW to be analyzed, CMS analysis is increasingly being performed completely outside of CMSSW. Your analysis group might have a framework that uses standalone ROOT, RDataFrame, or Scientific Python (e.g. numpy) instead. CMSSW is needed if your analysis needs additional variables not present in NanoAOD (for example, long-lived particle analysis often need RECO-level objects like tracker or calorimeter hits). You will also probably need to use CMSSW for detector, trigger, and/or POG work.
The framework goes hand-in-hand with the “Event Data Model” (EDM), which is how CMS represents events computationally. CMS saves events in several formats along the reconstruction chain, including RAW (data straight from detector), RECO (reconstruction performed), AOD (analysis object data=reduced RECO for analysis), MiniAOD (reduced AOD to fit in CMS’s disk space in Run 2), and NanoAOD (even further reduced MiniAOD). The upstream data formats are typically archived to tape storage, and must be loaded onto disk to be used. MiniAOD and NanoAOD are typically available on disk. We will learn more about finding data in the next exercise.
In this exercise, we will learn the basics of CMSSW, from setting up the software environment to running simple jobs.
Setting up CMSSW
Login to cmslpc as usual, and run the following commands to create a CMSSW environment. This will create a folder CMSSW_13_0_10_cmsdas, which contains several subfolders. The most important folder is CMSSW_13_0_10_cmsdas/src, which is where you put your code.
cd ~/nobackup/cmsdas
source /cvmfs/cms.cern.ch/cmsset_default.sh
scram project -n "CMSSW_13_0_10_cmsdas" CMSSW_13_0_10
Add
cmsset_default.shFor convenience, we suggest you edit your
~/.bash_profilefile to call thecmsset_default.shscript automatically upon login. Add the whole line to this script.
Beware of your software environment
Note that software environments are generally not cross-compatible! Namely, if you setup an LCG software release and CMSSW in the same session (or conda, etc.), things will break. For this exercise, make sure you have logged into a fresh session on cmslpc, so that the LCG environment from the previous exercise is not set up.
What does the
13_0_10mean?Note the release number,
13_0_10: CMSSW is a massive project that is under continuous development, so we define “releases” that corresponds to a fixed snapshot at some point in time.CMSSW_13_0_*is the release used for “NanoAODv12,” the https://gitlab.cern.ch/cms-nanoAOD/nanoaod-doc/-/wikis/Releases/NanoAODv12 for Run 3 analysis. The first number in series (13) indicates a major cycle, the second number (0) a major release with new features with respect to the preceeding release, and the third number (10) a release with minor updates and bug fixes to the preceeding release. TheCMSSW_13_0_Xseries is used for pp data taking in 2023. For full release schedule, see CMSSW release schedule.
Next, execute the following commands to setup the environment in your current shell session:
cd CMSSW_13_0_10_cmsdas/src
cmsenv
This will provide you with a number of commands and environment variables. For example $CMSSW_BASE is a handy variable that points to your CMSSW folder.
Question 5.1
The following command prints the location of your CMSSW area. Copy-and-paste the answer into the Google form 5.
echo $CMSSW_BASE
Question 5.2
CMSSW is connected to several external tools, for example the Pythia generator. The following command prints the version of Pythia connected to your current CMSSW release. Fill in the version number in the Google form 5.
scram tool info pythia8
Git
CMS makes extensive use of git for code management, and you will use it throughout CMSDAS (CMSSW itself is managed as a git repository, but it’s a rather complicated example, so we won’t talk about CMSSW+git here). Here, we will simply use git to download some code. First, if you don’t already have a github account, go back to the [setup instructions][lesson-setup] and follow the directions, including setting up the SSH keys.
Choose your username wisely
Choose your username wisely, it will appear on all your contributions to CMS code! In fact, even if you already have an account, if you have a username like
edgelord1337, consider either changing it or making a second account.
Once you have an account, run the following commands to configure git on cmslpc replacing everything including the [brackets], [Account] is your github account username:
git config --global user.name "[Your name]"
git config --global user.email [Email]
git config --global user.github [Account]
Next, run the following commands to “clone” a repository. Make sure not to skip the cd line, as the code has to end up in the correct folder structure.
cd $CMSSW_BASE/src
git clone git@github.com:FNALLPC/LearnCMSSW MyAnalysis/LearnCMSSW
If the
git clonefails, it’s possible your SSH key was not setup correctly. Double check the [setup instructions][lesson-setup], and head to Mattermost for help.
This will copy all the code in the repository to $CMSSW_BASE/src/MyAnalysis/LearnCMSSW. Feel free to glance through it.
Question 5.3: git repo info
When you cloned the repository, you not only downloaded the code, but also setup a local git repository connected to the remote repository. Use the following commands to print out the URL of the remote repository, from which you cloned the code:
cd $CMSSW_BASE/src/MyAnalysis/LearnCMSSW git remote -vCopy and paste the first line into the Google form.
Running a CMSSW job
Now that we have the source code, we have to compile it. Execute the following to compile the package using scram, CMSSW’s build tool:
cd $CMSSW_BASE/src
scram b
Multi-core build
scram baccept an argument-jto use more cores for the compilation. Don’t go above-j4, as overloading the cores will negatively impact other users on your cmslpc interactive node.
Finally, let’s actually run some code. CMSSW jobs are configured through python files. We will use $CMSSW_BASE/src/MyAnalysis/test/zpeak_cfg.py, which is a simple configuration file that loads the plugin at $CMSSW_BASE/src/MyAnalysis/LearnCMSSW/plugins/ZPeakAnalyzer.cc. The ZPeakAnalyzer processes some dimuon events in MiniAOD format and produces some histograms (a bit of an uncommon workflow, as it is typically more efficient to make histograms from NanoAOD or another slimmed-down format). Launch CMSSW with the following:
cd $CMSSW_BASE/src/MyAnalysis/LearnCMSSW/test
cmsRun zpeak_cfg.py
The job will take a minute to run, periodically updating you on the progress. When it’s done, you should see a file ZPeak.root. Let’s open it and plot the Z peak:
root -l ZPeak.root
[0] TH1D* dimuonMass = (TH1D*)_file0->Get("zpeak_analyzer/dimuonMass")
[1] dimuonMass->Draw()
Question 5.4
Using the stat box drawn along with the histogram, what is the mean dimuon mass? Write your answer in the Google form.
Key Points
CMSSW is CMS’s software framework for data processing.
The framework consists of lots of C++ modules, which are configured using python.
CMSSW jobs are launched using commands like
cmsRun myCfg.pyWe provide an example EDAnalyzer and cfg.py file for plotting a Z peak directly from a MiniAOD file.
Analyzing simple ROOT ntuples like NanoAOD does not need CMSSW!
CMSDAS Pre-Exercise 6: Using the grid
Overview
Teaching: 0 min
Exercises: 60 minQuestions
Objectives
Learn how to find CMS data on the grid
Launch a MC (Monte Carlo) generation job using CRAB
Launch a MiniAOD processing job using CRAB
Optional exercise
This exercise is not a prerequisite for registering for CMSDAS. We encourage all students to do this exercises anyways, as you will certainly need to use the grid for analysis at some point.
Questions
For this lesson, please submit your answers using CMSDAS@LPC2026 Google Form 6.
Before you start
In this set of exercises, we will learn how to do a full-scale analysis using the Worldwide LHC Computing Grid This set of exercises will take considerably longer than the others. Having your storage space set up may take several days, grid jobs can take a few days to run, and there can be problems. Although the actual effort for this exercise is only a few hours, you should set aside about a week to complete these exercises. (Note: you may or may not use CRAB during CMSDAS; however, to do a real CMS analysis, you will certainly need to use CRAB and/or the storage resources at /store/user/.)
If you encounter any problems with the exercise, please reach out on Mattermost or send an email to CMSDASATLPC@fnal.gov with a detailed description of your problem. Outside of CMSDAS, you can find help at the CRAB troubleshooting twiki or the Computing Tools forum on cms-talk.
Note
This section assumes that you have an account on the LPC computers at FNAL. How to get one is explained in the setup instructions. However, those familiar with the CERN computing environment and somewhat familiar with CRAB can answer all the questions running at CERN only. For CMSDAS, we recommend using a LPC account at FNAL and making sure you have write access to T3_US_FNALLPC. For T3_US_FNALLPC, you can get your EOS area mapped to your grid certificate by following these instructions to do a CMS Storage Space Request.
Later on, you can check with your university contact for Tier 2 or Tier 3 storage area. Once you are granted the write permission to the specified site, for later analysis you can use CRAB as the below exercise but store the output to your Tier 2 or Tier 3 storage area.
AGAIN: To perform this set of exercises, an LPC account, Grid Certificate, and CMS VO membership are required. You should already have these things, but if not, follow the setup instructions.
Finding data on the grid
In this exercise, we will learn to locate CMS data on the grid. CMS data is stored around the world at various T1, T2, and T3 computing sites. We will first learn how to use the Data Aggregation Service (DAS) to locate data (not to be confused with the data analysis school in which you are partaking!).
There are two ways to use DAS to find data: through a website or via the command line. First, let’s use the DAS website. You will be asked for your grid certificate, which you should have loaded into your browser. (Also note that there may be a security warning message, which you will need to bypass.) Enter the following into the form:
dataset dataset=/DYJetsToLL_M-50_TuneCP5_13TeV-madgraphMLM-pythia8/RunIISummer20UL18NanoAODv9-106X_upgrade2018_realistic_v16_L1v1-v1/NANOAODSIM
The first “dataset” indicates that you’re searching for whole data sets; the second “dataset=/DYJets…” is a filter, returning only this specific data set. The search results should show exactly one hit, along with several links for more info.
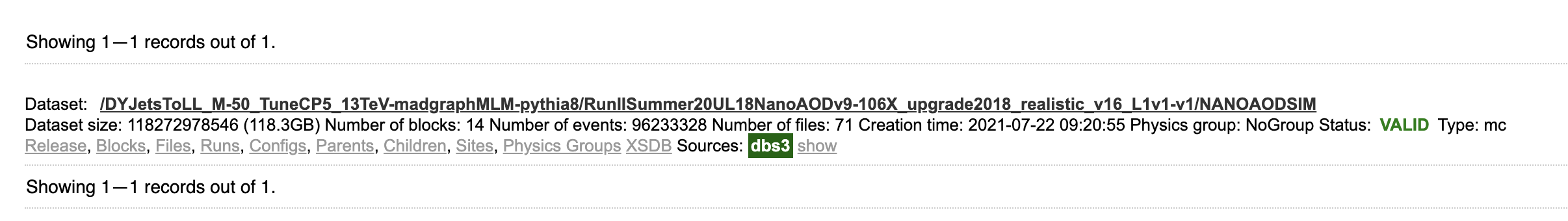
Let’s click on “Files,” which actually performs another search (you can see the exact search string in the search bar) for files in this data set. The results show 71 files, mostly around 2 GB in size with about 2 million events per file.
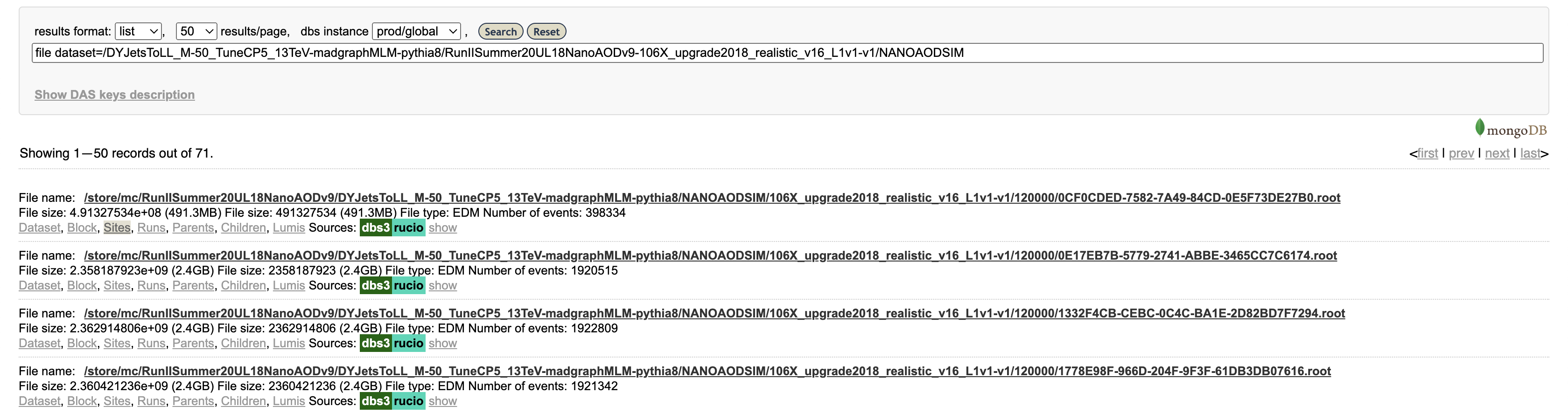
Finally, click on “Sites” for a single file. This will show the sites around the world that have this file available. This particular file is available at numerous sites, including T1_US_FNAL_Disk, T2_US_MIT, and T2_US_Purdue in the US.
More information about accessing data in the Data Aggregation Service can be found in WorkBookDataSamples.
CRAB introduction and prerequisites
Using DAS, we can locate our desired input data using, e.g., file dataset=/DYJetsToLL.... We could download all the files from the grid using xrootd. However, if you were to try this, you would quickly fill up your quota! Instead, we can use the CMS Remote Analysis Builder (CRAB). CRAB is a utility that distributes CMSSW jobs to the CMS grid (typically, but not always, using CPUs at the site where the input data is stored). The jobs will then transfer the reduced output (e.g., skimmed/slimmed ntuples or even histograms) to your /store/user/ space. CRAB is your entry point to the CMS’s significant CPU and storage resources around the world.
For general help or questions about CRAB, see the CRAB FAQ. The most recent CRAB tutorial can be found in the WorkBook under WorkBookCRABTutorial. This tutorial provides complete instructions for both beginner and expert CRAB users. We strongly recommend you to follow the CRAB tutorial after you finish these exercises.
Verify your grid certificate is OK
This exercise depends on obtaining a grid certificate and VOMS membership, but does not depend on any previous exercises.
After you’ve followed all the instructions above and installed your grid certificate, you need to verify it has all the information needed. Please install your grid certificate also on cmslpc-el9.fnal.gov
Login to cmslpc-el9.fnal.gov and initialize your proxy:
voms-proxy-init -voms cms
Then run the following command:
voms-proxy-info -all | grep -Ei "role|subject"
The response should look like this:
subject : /DC=ch/DC=cern/OU=Organic Units/OU=Users/CN=haweber/CN=713242/CN=Hannsjorg Weber/CN=2282089851
subject : /DC=ch/DC=cern/OU=Organic Units/OU=Users/CN=haweber/CN=713242/CN=Hannsjorg Weber
attribute : /cms/Role=NULL/Capability=NULL
attribute : /cms/uscms/Role=NULL/Capability=NULL
If you do not have the first attribute line listed above, you probably did not finish the VO registration in the setup. Double check this first, and otherwise reach out on Mattermost.
Question 6.1
Copy the output corresponding to the text in the output box above, using CMSDAS@LPC2026 Google Form 6.
Now that you have a valid grid proxy in the CMS VO, we will be able to run a command line client to check DAS to find CMS files and their information.
DAS client Question 6.2
Now let’s use the command line implementation of DAS to find some real collision data. This requires the valid CMS grid proxy. The
dasgoclientlets you query DAS just like the web interface. On cmslpc, enter the following query (along with agrepcommand to filter the result):dasgoclient -query="dataset dataset=/JetHT/Run2018C-UL2018_MiniAODv2_NanoAODv9-v1/NANOAOD" -json | grep "nevents"How many events are in this dataset? Enter the result in the Google form.
Obtain a /store/user area
This exercise pertains specifically to cmslpc. When you obtained your cmslpc account, you should have automatically been granted EOS storage at /store/user/YourUsername, however if your grid certificate was not provided or linked you may need to ensure that it is in Fermilab’s database. (As a refresher, you can review the instructions for using EOS at Using EOS at LPC.) You will configure your CRAB jobs to write output files to this space. However, it will not yet be linked to your grid certificate unless you provided it in the application form. If not linked, put in a Fermilab ServiceNow request following directions for CMS Storage Space Request. Note that this can take up to 1 business day (FNAL hours).
Setting up CRAB
Once you have a /store/user/ area connected with your grid certificate, we can start using CRAB.
In this exercise, you will generate a small MC sample yourself, and then publish it using DAS.
We will use CMSSW_13_0_13. Let’s setup a new CMSSW area for this exercise:
cd ~/nobackup/cmsdas/
scram project -n "CMSSW_13_0_13_mcgen" CMSSW_13_0_13
cd CMSSW_13_0_13_mcgen/src
cmsenv
CRAB is packaged along with CMSSW, so you should have it available now. Verify this by executing which crab; this should return something like /cvmfs/cms.cern.ch/common/crab.
Next, we will verify that your CRAB jobs have permission to write to your EOS storage. First, initialize your proxy:
voms-proxy-init -voms cms
Use the following command to check if CRAB has write access to your EOS area:
crab checkwrite --site= <site-name>
Specifically, for your cmslpc EOS storage, you should execute:
crab checkwrite --site=T3_US_FNALLPC
You should see a fair amount of output as CRAB tries several operations in your EOS storage space. At the end, hopefully you will see something like:
Will check write permission in the default location /store/user/<username>
Validating LFN /store/user/YourCERNUsername...
LFN /store/user/YourUsername is valid.
Will use `gfal-copy`, `gfal-rm` commands for checking write permissions
Will check write permission in /store/user/YourCERNUsername on site T3_US_FNALLPC
...
...
Success: Able to write in /store/user/YourCERNUsername on site T3_US_FNALLPC
Generate and publish a minimum-bias dataset with CRAB
In this exercise, we will generate a minimum bias Monte Carlo (MC) sample. A typical CMS MC job is configured with a “python fragment,” which is a small piece of python containing the necessary code to configure the generator inside CMSSW. Let’s download a recent minimum bias fragment:
cd $CMSSW_BASE/src
curl -s -k https://cms-pdmv-prod.web.cern.ch/mcm/public/restapi/requests/get_fragment/PPD-Run3Summer23GS-00003 --retry 3 --create-dirs -o Configuration/GenProduction/python/PPD-Run3Summer23GS-00003-fragment.py
The fragment contains the following lines; it mostly just sets up a special EDFilter module that runs Pythia inside CMSSW, along with a number of Pythia-specific settings:
import FWCore.ParameterSet.Config as cms
from Configuration.Generator.Pythia8CommonSettings_cfi import *
from Configuration.Generator.MCTunesRun3ECM13p6TeV.PythiaCP5Settings_cfi import *
generator = cms.EDFilter("Pythia8ConcurrentGeneratorFilter",
maxEventsToPrint = cms.untracked.int32(1),
pythiaPylistVerbosity = cms.untracked.int32(1),
filterEfficiency = cms.untracked.double(1.0),
pythiaHepMCVerbosity = cms.untracked.bool(False),
comEnergy = cms.double(13600.),
PythiaParameters = cms.PSet(
pythia8CommonSettingsBlock,
pythia8CP5SettingsBlock,
processParameters = cms.vstring(
'SoftQCD:inelastic = on'
),
parameterSets = cms.vstring('pythia8CommonSettings',
'pythia8CP5Settings',
'processParameters',
)
)
)
Next, we need to turn the fragment into a full-fledged CMSSW configuration file. We can do this with the helpful cmsDriver.py utility. Execute the following:
cd $CMSSW_BASE/src
scram b
cmsDriver.py Configuration/GenProduction/python/PPD-Run3Summer23GS-00003-fragment.py --python_filename cmsdas_minbias_cfg.py --eventcontent NANOAODGEN --step GEN,NANOGEN --datatier NANOAODSIM --customise Configuration/DataProcessing/Utils.addMonitoring --fileout file:cmsdas_minbias_nanogen.root --conditions 130X_mcRun3_2023_realistic_forMB_v1 --beamspot Realistic25ns13p6TeVEarly2023Collision --geometry DB:Extended --era Run3_2023 --no_exec --mc -n 20
Note: the scram b is necessary for CMSSW to be aware of the newly downloaded fragment file. This should have created a new file, cmsdas_minbias_cfg.py. Look over this file, and appreciate how much effort cmsDriver.py has saved us in creating this file automatically! This specific job will run 20 events through the GEN step (i.e. launching Pythia to generate truth-level events) and the NANOGEN step (which just dumps the truth particle info to a NanoAOD file); for simplicity, we are skipping the detector simulation.
Generating MC events locally
We want to test this Configuration file locally for a small number of events before we submit to CRAB for massive generation. To test this file, we can run
cmsRun cmsdas_minbias_cfg.py
This job will take a few minutes to run; you will see quite a lot of output from Pythia as it generates events. Once it finishes, you should have a brand new NANOGEN file, cmsdas_minbias_nanogen.root.
Question 6.3
What is the file size of
cmsdas_minbias_nanogen.root? Usels -lh.
Generate and publish MC dataset using CRAB
We usually need millions of MC events for a CMS analysis. The collaboration produces a lot of samples centrally, but you will occasionally need to make new samples for your analysis, or test MC. In this example, we will use CRAB to launch many minimum bias production jobs on the grid.
CRAB is configured using python configuration files (once again!). The complete documentation for these configuration files is here: CRAB3ConfigurationFile. For now, we have prepared a CRAB config for you; download it to cmslpc using:
cd $CMSSW_BASE/src
wget fnallpc.github.io/cms-das-pre-exercises/code/cmsdas_mc_crab.py
Below you also find the file:
Show/Hide
from WMCore.Configuration import Configuration config = Configuration() config.section_("General") config.General.requestName = 'cmsdas_minbias_test0' config.General.workArea = 'crabsubmit' config.section_("JobType") config.JobType.pluginName = 'PrivateMC' config.JobType.psetName = 'cmsdas_minbias_cfg.py' config.JobType.allowUndistributedCMSSW = True config.section_("Data") config.Data.outputPrimaryDataset = 'MinBias' config.Data.splitting = 'EventBased' config.Data.unitsPerJob = 20 NJOBS = 10 # This is not a configuration parameter, but an auxiliary variable that we use in the next line. config.Data.totalUnits = config.Data.unitsPerJob * NJOBS config.Data.publication = True config.Data.outputDatasetTag = 'MinBias_TuneCP5_13p6TeV-pythia8_cmsdas2025_test0' config.section_("Site") config.Site.storageSite = 'T3_US_FNALLPC'
To submit, simply execute the following:
crab submit -c cmsdas_mc_crab.py
(For the detail of the crab command, see CRABCommands.) You might be requested to enter your grid certificate password. You should get an output similar to this:
Will use CRAB configuration file cmsdas_mc_crab.py
Enter GRID pass phrase for this identity:
Importing CMSSW configuration cmsdas_minbias_cfg.py
Finished importing CMSSW configuration cmsdas_minbias_cfg.py
Sending the request to the server at cmsweb.cern.ch
Success: Your task has been delivered to the prod CRAB3 server.
Task name: 231110_212908:dryu_crab_cmsdas_minbias_test0
Project dir: crabsubmit/crab_cmsdas_minbias_test0
Please use ' crab status -d crabsubmit/crab_cmsdas_minbias_test0 ' to check how the submission process proceeds.
Log file is /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0/crab.log
Now you might notice a directory called crabsubmit is created under CMSSW_13_0_13_mcgen/src/. This directory contains all the essential metadata for your CRAB job, and is your main tool for interacting with your running jobs. Use crab status -d crabsubmit
IMPORTANT: do not modify or delete the files in this directory until you are 100% sure your job is finished! Your jobs will occasionally fail (for example, if the computing site is misconfigured, a network error occurs, the power goes out, …). You can use crab resubmit -d crabsubmit/the_job_dir to resubmit the jobs, but not if you have deleted the directory.
After you submitted the job successfully (give it a few moments), you can check the status of a task by executing the following CRAB command:
crab status -d crabsubmit/crab_cmsdas_minbias_test0
The crab status command will produce an output containing the task name, the status of the task as a whole, the details of how many jobs are in which state (submitted, running, transfering, finished, cooloff, etc.) and the location of the CRAB log (crab.log) file. It will also print the URLs of two web pages that one can use to monitor the jobs. In summary, it should look something like this:
Show/Hide
cmslpc178:~/nobackup/cmsdas/CMSSW_13_0_13_mcgen/src --> crab status -d crabsubmit/crab_cmsdas_minbias_test0 CRAB project directory: /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0 Task name: 231110_212908:username_crab_cmsdas_minbias_test0 Grid scheduler - Task Worker: crab3@vocms0198.cern.ch - crab-prod-tw01 Status on the CRAB server: SUBMITTED Task URL to use for HELP: https://cmsweb.cern.ch/crabserver/ui/task/231110_212908%3Ausername_crab_cmsdas_minbias_test0 Dashboard monitoring URL: https://monit-grafana.cern.ch/d/cmsTMDetail/cms-task-monitoring-task-view?orgId=11&var-user=username&var-task=231110_212908%3Ausername_crab_cmsdas_minbias_test0&from=1699648148000&to=now Status on the scheduler: SUBMITTED Jobs status: idle 100.0% (10/10) No publication information available yet Log file is /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0/crab.log
Now you can take a break and have some fun. Come back after couple hours or so and check the status again.
Show/Hide
[tonjes@cmslpc101 src]$ crab status crabsubmit/cmsdas_minbias_test0 CRAB project directory: /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0 Task name: 231110_212908:username_crab_cmsdas_minbias_test0 Grid scheduler - Task Worker: crab3@vocms0122.cern.ch - crab-prod-tw01 Status on the CRAB server: SUBMITTED Task URL to use for HELP: https://cmsweb.cern.ch/crabserver/ui/task/211024_214242%3Abelt_cmsdas_minbias_test0 Dashboard monitoring URL: https://monit-grafana.cern.ch/d/cmsTMDetail/cms-task-monitoring-task-view?orgId=11&var-user=belt&var-task=211024_214242%3Abelt_cmsdas_minbias_test0&from=1635108162000&to=now Status on the scheduler: COMPLETED Jobs status: finished 100.0% (10/10) Publication status of 1 dataset(s): done 100.0% (10/10) (from CRAB internal bookkeeping in transferdb) Output dataset: /MinBias/belt-CMSDAS2021_CRAB3_MC_generation_test0-67359df6f8a0ef3c567d7c8fea38a809/USER Output dataset DAS URL: https://cmsweb.cern.ch/das/request?input=%2FMinBias%2Fbelt-CMSDAS2021_CRAB3_MC_generation_test0-67359df6f8a0ef3c567d7c8fea38a809%2FUSER&instance=prod%2Fphys03 Warning: the max jobs runtime is less than 30% of the task requested value (1250 min), please consider to request a lower value for failed jobs (allowed through crab resubmit) and/or improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task. Warning: the average jobs CPU efficiency is less than 50%, please consider to improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task Summary of run jobs: * Memory: 39MB min, 84MB max, 43MB ave * Runtime: 0:04:55 min, 0:45:15 max, 0:08:59 ave * CPU eff: 7% min, 73% max, 22% ave * Waste: 1:15:29 (46% of total) Log file is /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0/crab.log
Note: If you specified T3_US_FNALLPC as your output directory, CRAB will write the output to your eos area. You can see them at something like eosls /store/user/username/MinBias/MinBias_TuneCP5_13p6TeV-pythia8_cmsdas2025_test0/....
From the bottom of the output, you can see the name of the dataset and the DAS link to it. Congratulations! This is the your first CMS dataset.
There is some magic going on under the hood here. For example, if you were to simply cmsRun cmsdas_minbias_cfg.py ten times, you would get 10 identical output files. CRAB takes care of assigning each job an independent seed for the random number generator, so that each file contains unique events!
Question 6.4
What is the dataset name you published?
Processing MiniAOD with CRAB
In this exercise, we will use CRAB to run the ZPeakAnalyzer module from the previous exercise. Remember that we were using a different CMSSW release, CMSSW_13_0_10_cmsdas, so make sure to logout, login, and setup the correct release:
cd ~/nobackup/cmsdas/CMSSW_13_0_10_cmsdas/src
cmsenv
Download another CRAB configuration file,
cd $CMSSW_BASE/src
wget fnallpc.github.io/cms-das-pre-exercises/code/cmsdas_zpeak_crab.py
The configuration file looks like this:
Show/Hide
import os from WMCore.Configuration import Configuration config = Configuration() config.section_("General") config.General.requestName = 'cmsdas_zpeak_test0' config.General.workArea = 'crabsubmit' config.section_("JobType") config.JobType.pluginName = 'Analysis' config.JobType.psetName = os.path.expandvars('$CMSSW_BASE/src/MyAnalysis/LearnCMSSW/test/zpeak_cfg.py') config.JobType.allowUndistributedCMSSW = True config.section_("Data") config.Data.inputDataset = '/DoubleMuon/Run2016C-03Feb2017-v1/MINIAOD' config.Data.inputDBS = 'global' config.Data.splitting = 'LumiBased' config.Data.unitsPerJob = 50 config.Data.lumiMask = 'https://cms-service-dqmdc.web.cern.ch/CAF/certification/Collisions16/13TeV/Cert_271036-275783_13TeV_PromptReco_Collisions16_JSON.txt' config.Data.runRange = '275776-275782' config.section_("Site") config.Site.storageSite = 'T3_US_FNALLPC'
Most of this file is similar to the previous MC generation job, but there are a few key differences. The runRange parameter is used to process only a small number of runs (to save time, effort, and CO2 in this exercise). The lumiMask parameter specifies which runs and lumisections are good for analysis (each detector is responsible for certifying which lumisections are good or bad; for example, HCAL would mark data as bad if the bias voltage for the photomultipliers was incorrect).
Run CRAB
Now go through the same process for this config file. You submit it with
crab submit -c cmsdas_zpeak_crab.py
and check the status with
crab status
After a while, you should see something like below:
Show/Hide
cmslpc154:~/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit --> crab status -d crab_cmsdas_zpeak_test0/ Your user certificate is going to expire in 1 days. See: https://twiki.cern.ch/twiki/bin/view/CMSPublic/WorkBookStartingGrid#ObtainingCert CRAB project directory: /uscms_data/d3/dryu/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit/crab_cmsdas_zpeak_test0 Task name: 231130_123342:dryu_crab_cmsdas_zpeak_test0 Grid scheduler - Task Worker: crab3@vocms0121.cern.ch - crab-prod-tw01 Status on the CRAB server: SUBMITTED Task URL to use for HELP: https://cmsweb.cern.ch/crabserver/ui/task/231130_123342%3Adryu_crab_cmsdas_zpeak_test0 Dashboard monitoring URL: https://monit-grafana.cern.ch/d/cmsTMDetail/cms-task-monitoring-task-view?orgId=11&var-user=dryu&var-task=231130_123342%3Adryu_crab_cmsdas_zpeak_test0&from=1701344022000&to=now Status on the scheduler: COMPLETED Jobs status: finished 100.0% (31/31) Output dataset: /FakeDataset/fakefile-FakePublish-93f81f35bfff96a473e04044a2f7a529/USER No publication information available yet Warning: the max jobs runtime is less than 30% of the task requested value (1250 min), please consider to request a lower value for failed jobs (allowed through crab resubmit) and/or improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task. Warning: the average jobs CPU efficiency is less than 50%, please consider to improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task Summary of run jobs: * Memory: 12MB min, 783MB max, 540MB ave * Runtime: 0:01:44 min, 0:04:03 max, 0:03:52 ave * CPU eff: 6% min, 63% max, 28% ave * Waste: 3:15:10 (62% of total) Log file is /uscms_data/d3/dryu/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit/crab_cmsdas_zpeak_test0/crab.log
Create reports of data analyzed
Once all jobs are finished (see crab status above), you can print a summary:
crab report
You’ll get something like this:
Show/Hide
Summary from jobs in status 'finished': Number of files processed: 64 Number of events read: 2167324 Number of events written in EDM files: 0 Number of events written in TFileService files: 0 Number of events written in other type of files: 0 Processed lumis written to processedLumis.json Summary from output datasets in DBS: Number of events: /FakeDataset/fakefile-FakePublish-93f81f35bfff96a473e04044a2f7a529/USER: 0 Additional report lumi files: Input dataset lumis (from DBS, at task submission time) written to inputDatasetLumis.json Lumis to process written to lumisToProcess.json Log file is /uscms_data/d3/dryu/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit/crab_cmsdas_zpeak_test0/crab.log
Question 6.5
How many events were read? Use
crab report -d crabsubmit/crab_cmsdas_zpeak_test0.
Optional: View the reconstructed Z peak in the combined data
Using CRAB, we have analyzed a lot more events than the local job in the previous exercise. Let’s plot the fruits of our labor. We can use the hadd command to combine the histograms from the several CRAB output files:
hadd ZPeak.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_1.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_10.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_11.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_13.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_14.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_15.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_16.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_17.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_18.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_19.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_2.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_20.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_21.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_22.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_25.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_26.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_27.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_28.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_29.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_3.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_30.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_31.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_4.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_5.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_6.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_7.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_8.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_9.root
Note: the author of this exercise did not actually type out this command by hand! Rather, I first obtained a list of files using eosls /store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/, and then used multiple cursors in Sublime Text/Visual Studio Code to turn the list into this long command. You could also generate the command programatically. Tricks like this often come in handy. Also note that the final file is less than 5GB in size. Many filesystems/networks do not do well with much larger files.
Open the new ZPeak.root file , and plot the histogram. You should have many more events than the last exercise!
Where to find more on CRAB
- CRAB Home
- CRAB FAQ
- CRAB troubleshooting guide: Steps to address the problems you experience with CRAB and how to ask for support.
- CMS Computing Tools mailing list, where to send feedback and ask support in case of jobs problem (please send to us your crab task HELP URL from crab status output).
Note also that all CMS members using the Grid subscribe to the Grid Annoucements CMS HyperNews forum. Important CRAB announcements will be announced on the CERN Computing Announcement HyperNews forum.
Last reviewed: 2024/10/28 by David Yu
Key Points
CMS data is stored around the world at various T1, T2, and T3 computing sites.
Use the Data Aggregation Service (DAS) to search for CMS data.
The CMS Remote Analysis Builder (CRAB) utility lets you launch jobs on the CMS grid.
Grid jobs run around the world, typically (but not always) using CPUs at the same site as the data.