CMS Data Analysis School Tracking and Vertexing Short Exercise - The five basic track variables
Overview
Teaching: 15 min
Exercises: 15 minQuestions
What are the main track variables used in CMS collaboration in different data formats?
How can we access them?
Which are the track quality variables?
How do the distributions of track variables before and after high purity track selection change?
Objectives
Being familiar with tracks collections and their variables.
Being aware of track information stored in different data format.
Accessing information and plot basic variables using ROOT.
One of the oldest tricks in particle physics is to put a track-measuring device in a strong, roughly uniform magnetic field so that the tracks curve with a radius proportional to their momenta (see derivation). Apart from energy loss and magnetic field inhomogeneities, the particles’ trajectories are helices. This allows us to measure a dynamic property (momentum) from a geometric property (radius of curvature).
A helical trajectory can be expressed by five parameters, but the parameterization is not unique. Given one parameterization, we can always re-express the same trajectory in another parameterization. Many of the data fields in a CMSSW reco::Track are alternate ways of expressing the same thing, and there are functions for changing the reference point from which the parameters are expressed. (For a much more detailed description, see this page.)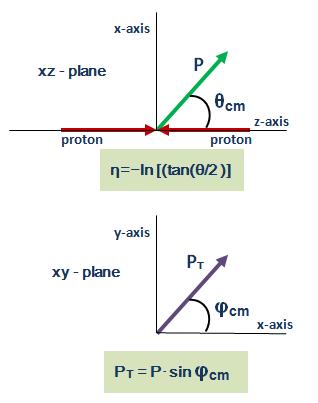
The five basic track parameters
signed radius of curvature (units of cm), which is proportional to particle charge divided by the transverse momentum, pT (units of GeV);
angle of the trajectory at a given point on the helix, in the plane transverse to the beamline (usually called φ);
angle of the trajectory at a given point on the helix with respect to the beamline (θ, or equivalently λ = π/2 - θ), which is usually expressed in terms of pseudorapidity (η = −ln(tan(θ/2)));
offset or impact parameter relative to some reference point (usually the beamspot or a selected primary vertex), in the plane transverse to the beamline (usually called dxy);
impact parameter relative to a reference point (beamspot or a selected primary vertex), along the beamline (usually called dz).
The exact definitions are given in the reco::TrackBase header file. This is also where most tracking variables and functions are defined. The rest are in the reco::Track header file, but most data fields in the latter are accessible only in RECO (full data record), not AOD/MiniAOD/NanoAOD (the subsets that are available to physics analyses).
Accessing track variables
Create print.py (for example emacs -nw print.py, or use your favorite text editor) in TrackingShortExercize/, then copy-paste the following code and run it (python3 print.py). Please note, if your run321457_ZeroBias_AOD.root is not in the directory you’re working from, be sure to use the appropriate path in line 2.
import DataFormats.FWLite as fwlite
events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
tracks = fwlite.Handle("std::vector<reco::Track>")
for i, event in enumerate(events):
if i >= 5: break # print info only about the first 5 events
print( "Event", i)
event.getByLabel("generalTracks", tracks)
numTotal = tracks.product().size()
print( "total tracks: ", numTotal)
for j, track in enumerate(tracks.product()):
if j >= 5: break # print info only about the first 5 tracks
print( " Track", j, end = " ")
print( "\t charge/pT: %.3f" %(track.charge()/track.pt()), end = " ")
print( "\t phi: %.3f" %track.phi(), end = " ")
print( "\t eta: %.3f" %track.eta(), end = " ")
print( "\t dxy: %.4f" %track.dxy(), end = " ")
print( "\t dz: %.4f" %track.dz())
to print all tracks comment out the line: if j >= 5: break as # if j >= 5: break
The first three lines load the FWLite framework, the .root data file, and prepare a Handle for the track collection using its full C++ name (std::vector). In each event, we load the tracks labeled generalTracks and loop over them, printing out the five basic track variables for each. The C++ equivalent of this is hidden below (longer and more difficult to write and compile, but faster to execute on large datasets) and is optional for this entire short exercise.
C++ version
First generate a template (also referred to as a skeleton) for analyzing an AOD root file with CMSSW with
mkedanlzr. Make the template inside theTrackingShortExercize/directory:cd ${CMSSW_BASE}/src/TrackingShortExercize mkedanlzr PrintOutTracksYou may see the files and directories that were produced with
mkedanlzrby running:tree PrintOutTracksoutput of the tree command
PrintOutTracks/ |-- plugins/ | |-- BuildFile.xml | `-- PrintOutTracks.cc |-- python `-- test |-- test_catch2_PrintOutTracks.cc |-- test_catch2_main.cc `-- BuildFile.xml Total: 3 directories, 5 filesNow, you may edit the template files to perform operations on the event data and produce output - such as histograms or print outs of diagnostic information to screen. ( To edit the files you may use your favorite text editor e.g.
emacs -nw PrintOutTracks/plugins/PrintOutTracks.xml):Inside the
PrintOutTracksclass definition (one line below the member data comment, before the};), replaceedm::EDGetTokenT<TrackCollection> tracksToken_;with:edm::EDGetTokenT<edm::View<reco::Track> > tracksToken_; //used to select which tracks to read from configuration file int indexEvent_;Inside the
PrintOutTracks::PrintOutTracks(const edm::ParameterSet& iConfig)constructor before the{, modify the consumes statement to read:: tracksToken_(consumes<edm::View<reco::Track> >(iConfig.getUntrackedParameter<edm::InputTag>("tracks", edm::InputTag("generalTracks")) ))after the
//now do what ever initialization is neededcomment:indexEvent_ = 0;Put the following inside the
PrintOutTracks::analyzemethod:// printf requires the package: #include <stdio.h>, but it is loaded elsewere in the framework // another useful method of displaying information is with std::cout printf("Event %i\n", indexEvent_); edm::Handle<edm::View<reco::Track> > trackHandle; iEvent.getByToken(tracksToken_, trackHandle); if ( !trackHandle.isValid() ) return; const edm::View<reco::Track>& tracks = *trackHandle; size_t iTrack = 0; for ( auto track : tracks ) { if(iTrack>=5){ iTrack++; continue;} printf( " Track %zu", iTrack); printf( "\t charge/pT: %.3f", (track.charge()/track.pt())); printf( "\t phi: %.3f", track.phi()); printf( "\t eta: %.3f", track.eta()); printf( "\t dxy: %.4f", track.dxy()); printf( "\t dz: %.4f\n" , track.dz()); iTrack++; } std::cout<<"total tracks: "<< iTrack<<std::endl; ++indexEvent_;Now compile it by running:
scram build -j 4if compilation fails, the following lines may need to be added:
at the top of
PrintOutTracks/plugins/BuildFile.xml<use name="DataFormats/TrackReco"/>and in
PrintOutTracks/plugins/PrintOutTracks.ccthe following in the#includesection:#include <iostream> #include "DataFormats/TrackReco/interface/Track.h" #include "DataFormats/TrackReco/interface/TrackFwd.h" #include "FWCore/Utilities/interface/InputTag.h"Go back to
TrackingShortExercize/and create a CMSSW configuration file namedrun_cfg.py(emacs -nw run_cfg.py):import FWCore.ParameterSet.Config as cms process = cms.Process("RUN") process.source = cms.Source("PoolSource", fileNames = cms.untracked.vstring("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")) process.maxEvents = cms.untracked.PSet(input = cms.untracked.int32(5)) process.MessageLogger = cms.Service("MessageLogger", destinations = cms.untracked.vstring("cout"), cout = cms.untracked.PSet(threshold = cms.untracked.string("ERROR"))) process.PrintOutTracks = cms.EDAnalyzer("PrintOutTracks") process.PrintOutTracks.tracks = cms.untracked.InputTag("generalTracks") process.path = cms.Path(process.PrintOutTracks)And, finally, run it with:
cmsRun run_cfg.pyThis will produce the same output as the Python script, but it can be used on huge datasets. Though the language is different, notice that C++ and
FWLiteuse the same names for member functions:charge(),pt(),phi(),eta(),dxy(), anddz(). That is intentional: you can learn what kinds of data are available with interactiveFWLiteand then use the same access methods when writing GRID jobs. There is another way to accessFWLitewith ROOT’s C++-like syntax.The plugin is here:
/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/MyDirectory/PrintOutTracks/plugins/PrintOutTracks.ccTherun_cfg.pyis here:/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/MyDirectory/PrintOutTracks/test/run_cfg.py
Track quality variables
The first thing you should notice is that each event has hundreds of tracks. That is because hadronic collisions produce large numbers of particles and generalTracks is the broadest collection of tracks identified by CMSSW reconstruction. Some of these tracks are not real (ghosts, duplicates, noise…) and a good analysis should define quality cuts to select tracks requiring a certain quality.
Some analyses remove spurious tracks by requiring them to come from the beamspot (small dxy, dz). Some require high-momentum (usually high transverse momentum, pT), but that would be a bad idea in a search for decays with a small mass difference such as ψ’ → J/ψ π+π-. In general, each analysis group should review their own needs and ask the Tracking POG about standard selections.
Some of these standard selections have been encoded into a quality flag with three categories: loose, tight, and highPurity. All tracks delivered to the analyzers are at least loose, tight is a subset of these that are more likely to be real, and highPurity is a subset of tight with even stricter requirements. There is a trade-off: loose tracks have high efficiency but also high backgrounds, highPurity has slightly lower efficiency but much lower backgrounds, and tight is in between (see also the plots below). As of CMSSW 7.4, these are all calculated using MVAs (MultiVariate Analysis techniques) for the various iterations. In addition to the status bits, it’s also possible to access the MVA values directly.
Create a new print.py (with a unique name like printMVA.py) file with the following lines:
Add a Handle to the MVA values:
import DataFormats.FWLite as fwlite
events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
tracks = fwlite.Handle("std::vector<reco::Track>")
MVAs = fwlite.Handle("std::vector<float>")
The event loop should be updated to this:
for i, event in enumerate(events):
if i >= 5: break # only the first 5 events
print( "Event", i)
event.getByLabel("generalTracks", tracks)
event.getByLabel("generalTracks", "MVAValues", MVAs)
numTotal = tracks.product().size()
if numTotal == 0: continue
numLoose = 0
numTight = 0
numHighPurity = 0
for j, (track, mva) in enumerate(zip(tracks.product(), MVAs.product())):
if track.quality(track.qualityByName("loose")): numLoose += 1
if track.quality(track.qualityByName("tight")): numTight += 1
if track.quality(track.qualityByName("highPurity")): numHighPurity += 1
if j<50 or 55<j: continue
print( " Track", j, end = " ")
print( "\t charge/pT: %.3f" %(track.charge()/track.pt()), end = " ")
print( "\t phi: %.3f" %track.phi(), end = " ")
print( "\t eta: %.3f" %track.eta(), end = " ")
print( "\t dxy: %.4f" %track.dxy(), end = " ")
print( "\t dz: %.4f" %track.dz(), end = " ")
print( "\t nHits: %s" %track.numberOfValidHits(), "(%s P+ %s S)"%(track.hitPattern().numberOfValidPixelHits(),track.hitPattern().numberOfValidStripHits()), end = " ")
print( "\t algo: %s" %track.algoName(), end = " ")
print( "\t mva: %.3f" %mva)
print( "Event", i, end = " ")
print( "numTotal:", numTotal, end = " ")
print( "numLoose:", numLoose, "(%.1f %%)"%(float(numLoose)/numTotal*100), end = " ")
print( "numTight:", numTight, "(%.1f %%)"%(float(numTight)/numTotal*100), end = " ")
print( "numHighPurity:", numHighPurity, "(%.1f %%)"%(float(numHighPurity)/numTotal*100))
print("total events: ", dir(events))
print("total events: ", type(events))
print("total events: ", events.size())
to print all tracks comment out this line: if j<50 or 55<j: continue as # if j<50 or 55<j: continue
The C++-equivalent is hidden below.
C++ version
Following up in the previous C++ example, the following lines can be added to the PrintOutTracks plugin (the
EDAnalyzer).To class declaration:
edm::EDGetTokenT<edm::View<float> > mvaValsToken_;To constructor:
// after tracksToken_(...) , mvaValsToken_( consumes<edm::View<float> >(iConfig.getUntrackedParameter<edm::InputTag>("mvaValues", edm::InputTag("generalTracks", "MVAValues")) ) )Change your
analyzemethod to:printf("Event %i\n", indexEvent_); edm::Handle<edm::View<reco::Track> > trackHandle; iEvent.getByToken(tracksToken_, trackHandle); if ( !trackHandle.isValid() ) return; const auto numTotal = trackHandle->size(); if (numTotal==0U){ indexEvent_++; return;} edm::Handle<edm::View<float> > trackMVAstoreHandle; iEvent.getByToken(mvaValsToken_,trackMVAstoreHandle); if ( !trackMVAstoreHandle.isValid() ) return; auto numLoose = 0; auto numTight = 0; auto numHighPurity = 0; const edm::View<reco::Track>& tracks = *trackHandle; size_t iTrack = 0; for ( auto track : tracks ) { if (track.quality(track.qualityByName("loose")) ) ++numLoose; if (track.quality(track.qualityByName("tight")) ) ++numTight; if (track.quality(track.qualityByName("highPurity"))) ++numHighPurity; if(iTrack<50 || 55<iTrack){ iTrack++; continue;} printf( "\t Track %zu", iTrack); printf( "\t charge/pT: %.3f", (track.charge()/track.pt())); printf( "\t phi: %.3f", track.phi()); printf( "\t eta: %.3f", track.eta()); printf( "\t dxy: %.4f", track.dxy()); printf( "\t dz: %.4f" , track.dz()); printf( "\t nHits: %i", track.numberOfValidHits()); printf( "(%i P+ %i S)", track.hitPattern().numberOfValidPixelHits(),track.hitPattern().numberOfValidStripHits()); printf( "\t algo: %s" , track.algoName().c_str()); printf( "\t mva: %.3f\n", trackMVAstoreHandle->at(iTrack)); iTrack++; } printf ( "Event %i ", indexEvent_); printf ( "numTotal: %i ", numTotal); printf ( "numLoose: %i ", numLoose); printf ( "(%.1f %%) ",(float(numLoose)/numTotal*100)); printf ( "numTight: %i ", numTight); printf ( "(%.1f %%) ",(float(numTight)/numTotal*100)); printf ( "numHighPurity: %i ", numHighPurity); printf ( "(%.1f %%)\n",(float(numHighPurity)/numTotal*100)); ++indexEvent_;Go back to
TrackingShortExercize/and edit the CMSSW configuration file namedrun_cfg.py(emacs -nw run_cfg.py), adding the following line:process.PrintOutTracks.mvaValues = cms.untracked.InputTag("generalTracks","MVAValues")Now run the
analyzer:cmsRun run_cfg.pyThe
plugincan be found in/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/PrintOutTracks_MVA.ccTheCMSSW configfile can be found in/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run_cfg_MVA.py
Question 1
Now prepare plots for the track variables discussed above, as in the example below (name this file
plot_track_quantities.pyand put it inTrackingShortExercize/). Compare the distributions of track-quality-related variables (number of pixel hits, track goodness of fit, …, which are given in input to MVA classifiers) between tracks passing thehighPurityandLoosequality flags. Do these distributions make sense to you?
Answer
import DataFormats.FWLite as fwlite import ROOT from ROOT import gROOT import os events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root") tracks = fwlite.Handle("std::vector<reco::Track>") hist_pt = ROOT.TH1F("pt", "track pt; p_{T} [GeV]", 100, 0.0, 100.0) hist_eta = ROOT.TH1F("eta", "track eta; #eta", 60, -3.0, 3.0) hist_phi = ROOT.TH1F("phi", "track phi; #phi", 64, -3.2, 3.2) hist_loose_normChi2 = ROOT.TH1F("hist_loose_normChi2" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_loose_numPixelHits = ROOT.TH1F("hist_loose_numPixelHits", "pixel hits; # pixel hits" , 15, 0.0, 15.0) hist_loose_numValidHits = ROOT.TH1F("hist_loose_numValidHits", "valid hits; # valid hits" , 35, 0.0, 35.0) hist_loose_numTkLayers = ROOT.TH1F("hist_loose_numTkLayers" , "valid layers; # valid Tk layers" , 25, 0.0, 25.0) hist_highP_normChi2 = ROOT.TH1F("hist_highP_normChi2" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_highP_numPixelHits = ROOT.TH1F("hist_highP_numPixelHits", "pixel hits; # pixel hits" , 15, 0.0, 15.0) hist_highP_numValidHits = ROOT.TH1F("hist_highP_numValidHits", "valid hits; # valid hits" , 35, 0.0, 35.0) hist_highP_numTkLayers = ROOT.TH1F("hist_highP_numTkLayers" , "valid layers; # valid Tk layers" , 25, 0.0, 25.0) hist_highP_normChi2.SetLineColor(ROOT.kRed) hist_highP_numPixelHits.SetLineColor(ROOT.kRed) hist_highP_numValidHits.SetLineColor(ROOT.kRed) hist_highP_numTkLayers.SetLineColor(ROOT.kRed) for i, event in enumerate(events): event.getByLabel("generalTracks", tracks) for track in tracks.product(): hist_pt.Fill(track.pt()) hist_eta.Fill(track.eta()) hist_phi.Fill(track.phi()) if track.quality(track.qualityByName("loose")): hist_loose_normChi2 .Fill(track.normalizedChi2()) hist_loose_numPixelHits.Fill(track.hitPattern().numberOfValidPixelHits()) hist_loose_numValidHits.Fill(track.hitPattern().numberOfValidHits()) hist_loose_numTkLayers .Fill(track.hitPattern().trackerLayersWithMeasurement()) if track.quality(track.qualityByName("highPurity")): hist_highP_normChi2 .Fill(track.normalizedChi2()) hist_highP_numPixelHits.Fill(track.hitPattern().numberOfValidPixelHits()) hist_highP_numValidHits.Fill(track.hitPattern().numberOfValidHits()) hist_highP_numTkLayers .Fill(track.hitPattern().trackerLayersWithMeasurement()) if i > 500: break gROOT.SetBatch(True) # this allows pyroot to run in batch mode - which prevents the histograms from being displayed every time they are drawn. c = ROOT.TCanvas( "c", "c", 800, 800) # make an output directory odir = "{0}/{1}/".format("plots", "highP") if not os.path.isdir(odir): os.makedirs(odir) # draw and save histograms as pdf files (can alternatively save as png by replacing .pdf with .png hist_pt.Draw() c.SetLogy() c.SaveAs(odir+"track_pt.pdf") c.SetLogy(False) hist_eta.Draw() c.SaveAs(odir+"track_eta.pdf") hist_phi.Draw() c.SaveAs(odir+"track_phi.pdf") hist_highP_normChi2.DrawNormalized() hist_loose_normChi2.DrawNormalized('same') c.SaveAs(odir+"track_normChi2.pdf") hist_highP_numPixelHits.DrawNormalized() hist_loose_numPixelHits.DrawNormalized('same') c.SaveAs(odir+"track_nPixelHits.pdf") hist_highP_numValidHits.DrawNormalized() hist_loose_numValidHits.DrawNormalized('same') c.SaveAs(odir+"track_nValHits.pdf") hist_highP_numTkLayers.DrawNormalized() hist_loose_numTkLayers.DrawNormalized('same') c.SaveAs(odir+"track_nTkLayers.pdf")Optional: save histograms to root file
Additional lines of code to create a root file and save the histograms there
this way you can dynamically view and style histograms (and save to pdf/png with another python script) without having to loop over the AOD/miniAOD everytime you want to change histogram or canvas attributes (like color, line size, labels etc.). Plots are most useful when they clearly describe the information being presented. This technique is also useful for creating “ratio” plots and “pull” plots.
# OPTIONAL: create a root file and save the histograms there ofile = ROOT.TFile.Open(odir+"hists.root", "RECREATE") ofile.WriteObject(hist_pt, "hist_pt") ofile.WriteObject(hist_eta, "hist_eta") ofile.WriteObject(hist_phi, "hist_phi") ofile.WriteObject(hist_loose_normChi2, "hist_loose_normChi2") ofile.WriteObject(hist_highP_normChi2, "hist_highP_normChi2") ofile.WriteObject(hist_loose_numPixelHits, "hist_loose_numPixelHits") ofile.WriteObject(hist_highP_numPixelHits, "hist_highP_numPixelHits") ofile.WriteObject(hist_loose_numValidHits, "hist_loose_numValidHits") ofile.WriteObject(hist_highP_numValidHits, "hist_highP_numValidHits") ofile.WriteObject(hist_loose_numTkLayers, "hist_loose_numTkLayers") ofile.WriteObject(hist_highP_numTkLayers, "hist_highP_numTkLayers")Optional: If having trouble viewing the histograms
You may try displaying the files directly from cmslpc or lxplus using the
displaycommand:display plots/highP/track_pt.pdfthis requires all display settings to the remote connection to be correctly configured.
Sometimes it can be slow to view histograms on cmslpc or lxplus. This can happen because of a slow network connection to the remote computers - possibly when connected far away from Fermilab or CERN.
One option to view histograms is to copy them to the local machine you are working from (i.e. laptop) - this can be done with commands like
scporrsync. You may find more information on these commands by looking at the manuals. From the command line runman scporman rsync. You can also google these command to find more documentation and examples.The basic syntax of both commands (for copying a file from a remote machine to the local machine) is:
<scp/rsync> <options/flags> <source-path> <destination-path>where you must replace the text for each
< >with the relavent information.Recommending to copy files with scp for now (it is faster to learn): First make a directory on the your local computer with
mkdir histograms. And relocate to that dirctory withcd histogramsmkdir histograms cd histograms
- Determine the absolute path on cmslpc of the directory (or file) you wish to have on the local computer:
pwdon cmslpc.- use your cmslpc/lxplus username (if it is the same as on the local machine then ${USER} will work)
- if you set up an ssh config file on the local machine - you can use the hostname from that configuration
The following examples may help guide you to the right command (replace items surrounded by
< >):scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<cmslpc-ssh-hostname>:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:/uscms/homes/<first-letter-of-lpc-username>/<lpc-username>/nobackup/CMSSW_10_6_30_patch1_cmsdas/src/TrackingShortExercize/plots/highP" ./ scp -rp "$USER@cmslpc-el8.fnal.gov:/uscms/homes/${USER:0:1}/${USER}/nobackup/CMSSW_10_6_30_patch1_cmsdas/src/TrackingShortExercize/plots/highP" ./When you have time
rsyncis worth learning about. Note: a main difference betweenscpandrsyncis:scpwill copy all items from the remote machine andrsyncwill only copy the files that don’t exist locally and those that have been updated. Thersynccommand can be very useful - because it can be re-run multiple times without wasting time copying things that are already up to date. The caveat is that you must pay attention to the/at the the end of the path names if you are copying directories for both the . With the/means contents of directory, without the/is including the directory name. This must be considered for both the source path and the destination path.
Track information in MiniAOD
There is no track collection stored in MiniAOD analogous to the generalTracks in AOD. Tracks associated to charged Particle Flow CandidatesPFCandidates are accessible directly from the packedPFCandidates collection for tracks with pT > 0.5 GeV. For tracks between 0.5 and 0.95 GeV the track information is stored with reduced precision. Tracks not associated with PF candidates are in the lostTracks collection if their pT is above 0.95 GeV or they are associated with a secondary vertex or a KS or Lambda candidate. However, for both the tracks associated to the PFCandidates and the lostTracks, the highQuality track selection is used. Tracks with lower quality are not available in MiniAOD at all. In addition, tracks in the lostTracks collection are required to have at least 8 hits of which at least one has to be a pixel hit.
In particular, the track information saved in the PFCandidates is the following:
Track information in PFCandidates collection
- the uncertainty on the impact parameter
dzError(),dxyError()- the number of layers with hits on the track
- the sub/det and layer of first hit of the track
- the
reco::Trackof the candidate is provided by thepseudoTrack()method, with the following information stored:
- the pT,eta and phi of the original track (if those are different from the one of the original
PFCandidate)- an approximate covariance matrix of the track state at the vertex
- approximate
hitPattern()andtrackerExpectedHitsInner()that yield the correct number of hits, pixel hits, layers and the information returned bylostInnerHits()- the track normalized chisquare (truncated to an integer)
- the
highPurityquality flag set, if the original track had it.
Consider that the packedPFCandidates collects both charged and neutral candidates, therefore before trying to access the track information it is important to ensure that the candidate is charged and has the track information correctly stored (track.hasTrackDetails()).
Question 2
Write a simple script
print-comparison.pythat reads a MiniAOD file and the AOD file and compare plots of the same variables we looked at before forHighPuritytracks. For the track pT distributuon, focus on the low pT region below 5 GeV. Can you see any (non-statistical) difference with the previosu plots? The MiniAOD file is located here:root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_MINIAOD.root
Answer
import DataFormats.FWLite as fwlite import ROOT import os ROOT.gROOT.SetBatch(True) # this allows pyroot to run in batch mode - which prevents the histograms from being displayed every time they are drawn. events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_MINIAOD.root") eventsAOD = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root") tracks = fwlite.Handle("std::vector<pat::PackedCandidate>") losttracks = fwlite.Handle("std::vector<pat::PackedCandidate>") tracksAOD = fwlite.Handle("std::vector<reco::Track>") hist_pt = ROOT.TH1F("pt", "track pt; p_{T} [GeV]", 100, 0.0, 100.0) hist_lowPt = ROOT.TH1F("lowPt", "track pt; p_{T} [GeV]", 100, 0.0, 5.0) hist_eta = ROOT.TH1F("eta", "track eta; #eta", 60, -3.0, 3.0) hist_phi = ROOT.TH1F("phi", "track phi; #phi", 64, -3.2, 3.2) hist_normChi2 = ROOT.TH1F("hist_normChi2" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_numPixelHits = ROOT.TH1F("hist_numPixelHits", "pixel hits; # pixel hits" , 15, -0.5, 14.5) hist_numValidHits = ROOT.TH1F("hist_numValidHits", "valid hits; # valid hits" , 35, -0.5, 34.5) hist_numTkLayers = ROOT.TH1F("hist_numTkLayers" , "valid layers; # valid Tk layers" , 25, -0.5, 24.5) hist_pt_AOD = ROOT.TH1F("ptAOD", "track pt; p_{T} [GeV]", 100, 0.0, 100.0) hist_lowPt_AOD = ROOT.TH1F("lowPtAOD", "track pt; p_{T} [GeV]", 100, 0.0, 5.0) hist_eta_AOD = ROOT.TH1F("etaAOD", "track eta; #eta", 60, -3.0, 3.0) hist_phi_AOD = ROOT.TH1F("phiAOD", "track phi; #phi", 64, -3.2, 3.2) hist_normChi2_AOD = ROOT.TH1F("hist_normChi2AOD" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_numPixelHits_AOD = ROOT.TH1F("hist_numPixelHitsAOD", "pixel hits; # pixel hits" , 15, -0.5, 14.5) hist_numValidHits_AOD = ROOT.TH1F("hist_numValidHitsAOD", "valid hits; # valid hits" , 35, -0.5, 34.5) hist_numTkLayers_AOD = ROOT.TH1F("hist_numTkLayersAOD" , "valid layers; # valid Tk layers" , 25, -0.5, 24.5) hist_pt_AOD.SetLineColor(ROOT.kRed) hist_lowPt_AOD.SetLineColor(ROOT.kRed) hist_eta_AOD.SetLineColor(ROOT.kRed) hist_phi_AOD.SetLineColor(ROOT.kRed) hist_normChi2_AOD.SetLineColor(ROOT.kRed) hist_numPixelHits_AOD.SetLineColor(ROOT.kRed) hist_numValidHits_AOD.SetLineColor(ROOT.kRed) hist_numTkLayers_AOD.SetLineColor(ROOT.kRed) for i, event in enumerate(events): event.getByLabel("packedPFCandidates", "", tracks) event.getByLabel("lostTracks", "", losttracks) alltracks = [track for track in tracks.product()] alltracks += [track for track in losttracks.product()] for track in alltracks : if (not track.hasTrackDetails() or track.charge() == 0 ): continue if not track.trackHighPurity(): continue hist_pt.Fill(track.pt()) hist_lowPt.Fill(track.pt()) hist_eta.Fill(track.eta()) hist_phi.Fill(track.phi()) hist_normChi2 .Fill(track.pseudoTrack().normalizedChi2()) hist_numPixelHits.Fill(track.numberOfPixelHits()) hist_numValidHits.Fill(track.pseudoTrack().hitPattern().numberOfValidHits()) hist_numTkLayers .Fill(track.pseudoTrack().hitPattern().trackerLayersWithMeasurement()) if i > 1000: break for i, event in enumerate(eventsAOD): event.getByLabel("generalTracks", tracksAOD) for j, track in enumerate(tracksAOD.product()) : if not track.quality(track.qualityByName("highPurity")): continue hist_pt_AOD.Fill(track.pt()) hist_lowPt_AOD.Fill(track.pt()) hist_eta_AOD.Fill(track.eta()) hist_phi_AOD.Fill(track.phi()) hist_normChi2_AOD .Fill(track.normalizedChi2()) hist_numPixelHits_AOD.Fill(track.hitPattern().numberOfValidPixelHits()) hist_numValidHits_AOD.Fill(track.hitPattern().numberOfValidHits()) hist_numTkLayers_AOD .Fill(track.hitPattern().trackerLayersWithMeasurement()) if i > 1000: break c = ROOT.TCanvas( "c", "c", 800, 800) # make an output directory odir = "{0}/{1}/".format("plots", "miniAOD") if not os.path.isdir(odir): os.makedirs(odir) # draw and save histograms as pdf files (can alternatively save as png by replacing .pdf with .png hist_pt.Draw() hist_pt_AOD.Draw("same") c.SetLogy() c.SaveAs(odir+"track_pt_miniaod.pdf") hist_lowPt_AOD.Draw() hist_lowPt.Draw("same") c.SetLogy() c.SaveAs(odir+"track_lowPt_miniaod.pdf") c.SetLogy(False) hist_eta_AOD.Draw() hist_eta.Draw("same") c.SaveAs(odir+"track_eta_miniaod.pdf") hist_phi_AOD.Draw() hist_phi.Draw("same") c.SaveAs(odir+"track_phi_miniaod.pdf") hist_normChi2_AOD.Draw() hist_normChi2.Draw("same") c.SaveAs(odir+"track_normChi2_miniaod.pdf") hist_numPixelHits_AOD.Draw() hist_numPixelHits.Draw("same") c.SaveAs(odir+"track_nPixelHits_miniaod.pdf") hist_numValidHits_AOD.Draw() hist_numValidHits.Draw("same") c.SaveAs(odir+"track_nValHits_miniaod.pdf") hist_numTkLayers_AOD.Draw() hist_numTkLayers.Draw("same") c.SaveAs(odir+"track_nTkLayers_miniaod.pdf")Optional: save histograms to root file
see previus example for why you might want to do this.
# OPTIONAL: create a root file and save the histograms there ofile = ROOT.TFile.Open(odir+"hists_AOD.root", "RECREATE") ofile.WriteObject(hist_pt, "hist_pt") ofile.WriteObject(hist_pt_AOD, "hist_pt_AOD") ofile.WriteObject(hist_lowPt, "hist_lowPt") ofile.WriteObject(hist_lowPt_AOD, "hist_lowPt_AOD") ofile.WriteObject(hist_eta, "hist_eta") ofile.WriteObject(hist_eta_AOD, "hist_eta_AOD") ofile.WriteObject(hist_phi, "hist_phi") ofile.WriteObject(hist_phi_AOD, "hist_phi_AOD") ofile.WriteObject(hist_normChi2, "hist_normChi2") ofile.WriteObject(hist_normChi2_AOD, "hist_normChi2_AOD") ofile.WriteObject(hist_numPixelHits, "hist_numPixelHits") ofile.WriteObject(hist_numPixelHits_AOD, "hist_numPixelHits_AOD") ofile.WriteObject(hist_numValidHits, "hist_numValidHits") ofile.WriteObject(hist_numValidHits_AOD, "hist_numValidHits_AOD") ofile.WriteObject(hist_numTkLayers, "hist_numTkLayers") ofile.WriteObject(hist_numTkLayers_AOD, "hist_numTkLayers_AOD")Optional: copy histograms to local computer
Recommending to copy files with scp for now (it is faster to learn): First make a directory on the your local computer with
mkdir histograms. And relocate to that dirctory withcd histogramsmkdir histograms cd histograms
- Determine the absolute path on cmslpc of the directory (or file) you wish to have on the local computer:
pwdon cmslpc.- use your cmslpc/lxplus username (if it is the same as on the local machine then ${USER} will work)
- if you set up an ssh config file on the local machine - you can use the hostname from that configuration
The following examples may help guide you to the right command (replace items surrounded by
< >):scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<cmslpc-ssh-hostname>:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:/uscms/homes/<first-letter-of-lpc-username>/<lpc-username>/nobackup/CMSSW_14_0_19/src/TrackingShortExercize/plots/highP" ./ scp -rp "$USER@cmslpc-el8.fnal.gov:/uscms/homes/${USER:0:1}/${USER}/nobackup/CMSSW_14_0_19/src/TrackingShortExercize/plots/highP" ./When you have time
rsyncis worth learning about.
Key Points
The pre-selection of tracks to
HighPurityin MiniAOD also affects the distribution of the track quality parameters.All tracks are stored in the
generalTrackscollection in AOD.In MiniAOD they are accessible in a less straightforward way (
packedPFCandidates,lostTrackscollection) and not all tracks are available!