CMS Data Analysis School Tracking and Vertexing Short Exercise - The five basic track variables
Overview
Teaching: 15 min
Exercises: 15 minQuestions
What are the main track variables used in CMS collaboration in different data formats?
How can we access them?
Which are the track quality variables?
How do the distributions of track variables before and after high purity track selection change?
Objectives
Being familiar with tracks collections and their variables.
Being aware of track information stored in different data format.
Accessing information and plot basic variables using ROOT.
One of the oldest tricks in particle physics is to put a track-measuring device in a strong, roughly uniform magnetic field so that the tracks curve with a radius proportional to their momenta (see derivation). Apart from energy loss and magnetic field inhomogeneities, the particles’ trajectories are helices. This allows us to measure a dynamic property (momentum) from a geometric property (radius of curvature).
A helical trajectory can be expressed by five parameters, but the parameterization is not unique. Given one parameterization, we can always re-express the same trajectory in another parameterization. Many of the data fields in a CMSSW reco::Track are alternate ways of expressing the same thing, and there are functions for changing the reference point from which the parameters are expressed. (For a much more detailed description, see this page.)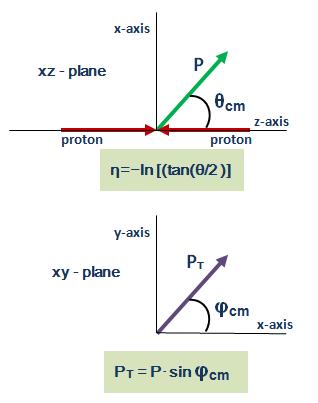
The five basic track parameters
signed radius of curvature (units of cm), which is proportional to particle charge divided by the transverse momentum, pT (units of GeV);
angle of the trajectory at a given point on the helix, in the plane transverse to the beamline (usually called φ);
angle of the trajectory at a given point on the helix with respect to the beamline (θ, or equivalently λ = π/2 - θ), which is usually expressed in terms of pseudorapidity (η = −ln(tan(θ/2)));
offset or impact parameter relative to some reference point (usually the beamspot or a selected primary vertex), in the plane transverse to the beamline (usually called dxy);
impact parameter relative to a reference point (beamspot or a selected primary vertex), along the beamline (usually called dz).
The exact definitions are given in the reco::TrackBase header file. This is also where most tracking variables and functions are defined. The rest are in the reco::Track header file, but most data fields in the latter are accessible only in RECO (full data record), not AOD/MiniAOD/NanoAOD (the subsets that are available to physics analyses).
Accessing track variables
Create print.py (for example emacs -nw print.py, or use your favorite text editor) in TrackingShortExercize/, then copy-paste the following code and run it (python3 print.py). Please note, if your run321457_ZeroBias_AOD.root is not in the directory you’re working from, be sure to use the appropriate path in line 2.
import DataFormats.FWLite as fwlite
events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
tracks = fwlite.Handle("std::vector<reco::Track>")
for i, event in enumerate(events):
if i >= 5: break # print info only about the first 5 events
print( "Event", i)
event.getByLabel("generalTracks", tracks)
numTotal = tracks.product().size()
print( "total tracks: ", numTotal)
for j, track in enumerate(tracks.product()):
if j >= 5: break # print info only about the first 5 tracks
print( " Track", j, end = " ")
print( "\t charge/pT: %.3f" %(track.charge()/track.pt()), end = " ")
print( "\t phi: %.3f" %track.phi(), end = " ")
print( "\t eta: %.3f" %track.eta(), end = " ")
print( "\t dxy: %.4f" %track.dxy(), end = " ")
print( "\t dz: %.4f" %track.dz())
to print all tracks comment out the line: if j >= 5: break as # if j >= 5: break
The first three lines load the FWLite framework, the .root data file, and prepare a Handle for the track collection using its full C++ name (std::vector). In each event, we load the tracks labeled generalTracks and loop over them, printing out the five basic track variables for each. The C++ equivalent of this is hidden below (longer and more difficult to write and compile, but faster to execute on large datasets) and is optional for this entire short exercise.
C++ version
First generate a template (also referred to as a skeleton) for analyzing an AOD root file with CMSSW with
mkedanlzr. Make the template inside theTrackingShortExercize/directory:cd ${CMSSW_BASE}/src/TrackingShortExercize mkedanlzr PrintOutTracksYou may see the files and directories that were produced with
mkedanlzrby running:tree PrintOutTracksoutput of the tree command
PrintOutTracks/ |-- plugins/ | |-- BuildFile.xml | `-- PrintOutTracks.cc |-- python `-- test |-- test_catch2_PrintOutTracks.cc |-- test_catch2_main.cc `-- BuildFile.xml Total: 3 directories, 5 filesNow, you may edit the template files to perform operations on the event data and produce output - such as histograms or print outs of diagnostic information to screen. ( To edit the files you may use your favorite text editor e.g.
emacs -nw PrintOutTracks/plugins/PrintOutTracks.xml):Inside the
PrintOutTracksclass definition (one line below the member data comment, before the};), replaceedm::EDGetTokenT<TrackCollection> tracksToken_;with:edm::EDGetTokenT<edm::View<reco::Track> > tracksToken_; //used to select which tracks to read from configuration file int indexEvent_;Inside the
PrintOutTracks::PrintOutTracks(const edm::ParameterSet& iConfig)constructor before the{, modify the consumes statement to read:: tracksToken_(consumes<edm::View<reco::Track> >(iConfig.getUntrackedParameter<edm::InputTag>("tracks", edm::InputTag("generalTracks")) ))after the
//now do what ever initialization is neededcomment:indexEvent_ = 0;Put the following inside the
PrintOutTracks::analyzemethod:// printf requires the package: #include <stdio.h>, but it is loaded elsewere in the framework // another useful method of displaying information is with std::cout printf("Event %i\n", indexEvent_); edm::Handle<edm::View<reco::Track> > trackHandle; iEvent.getByToken(tracksToken_, trackHandle); if ( !trackHandle.isValid() ) return; const edm::View<reco::Track>& tracks = *trackHandle; size_t iTrack = 0; for ( auto track : tracks ) { if(iTrack>=5){ iTrack++; continue;} printf( " Track %zu", iTrack); printf( "\t charge/pT: %.3f", (track.charge()/track.pt())); printf( "\t phi: %.3f", track.phi()); printf( "\t eta: %.3f", track.eta()); printf( "\t dxy: %.4f", track.dxy()); printf( "\t dz: %.4f\n" , track.dz()); iTrack++; } std::cout<<"total tracks: "<< iTrack<<std::endl; ++indexEvent_;Now compile it by running:
scram build -j 4if compilation fails, the following lines may need to be added:
at the top of
PrintOutTracks/plugins/BuildFile.xml<use name="DataFormats/TrackReco"/>and in
PrintOutTracks/plugins/PrintOutTracks.ccthe following in the#includesection:#include <iostream> #include "DataFormats/TrackReco/interface/Track.h" #include "DataFormats/TrackReco/interface/TrackFwd.h" #include "FWCore/Utilities/interface/InputTag.h"Go back to
TrackingShortExercize/and create a CMSSW configuration file namedrun_cfg.py(emacs -nw run_cfg.py):import FWCore.ParameterSet.Config as cms process = cms.Process("RUN") process.source = cms.Source("PoolSource", fileNames = cms.untracked.vstring("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")) process.maxEvents = cms.untracked.PSet(input = cms.untracked.int32(5)) process.MessageLogger = cms.Service("MessageLogger", destinations = cms.untracked.vstring("cout"), cout = cms.untracked.PSet(threshold = cms.untracked.string("ERROR"))) process.PrintOutTracks = cms.EDAnalyzer("PrintOutTracks") process.PrintOutTracks.tracks = cms.untracked.InputTag("generalTracks") process.path = cms.Path(process.PrintOutTracks)And, finally, run it with:
cmsRun run_cfg.pyThis will produce the same output as the Python script, but it can be used on huge datasets. Though the language is different, notice that C++ and
FWLiteuse the same names for member functions:charge(),pt(),phi(),eta(),dxy(), anddz(). That is intentional: you can learn what kinds of data are available with interactiveFWLiteand then use the same access methods when writing GRID jobs. There is another way to accessFWLitewith ROOT’s C++-like syntax.The plugin is here:
/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/MyDirectory/PrintOutTracks/plugins/PrintOutTracks.ccTherun_cfg.pyis here:/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/MyDirectory/PrintOutTracks/test/run_cfg.py
Track quality variables
The first thing you should notice is that each event has hundreds of tracks. That is because hadronic collisions produce large numbers of particles and generalTracks is the broadest collection of tracks identified by CMSSW reconstruction. Some of these tracks are not real (ghosts, duplicates, noise…) and a good analysis should define quality cuts to select tracks requiring a certain quality.
Some analyses remove spurious tracks by requiring them to come from the beamspot (small dxy, dz). Some require high-momentum (usually high transverse momentum, pT), but that would be a bad idea in a search for decays with a small mass difference such as ψ’ → J/ψ π+π-. In general, each analysis group should review their own needs and ask the Tracking POG about standard selections.
Some of these standard selections have been encoded into a quality flag with three categories: loose, tight, and highPurity. All tracks delivered to the analyzers are at least loose, tight is a subset of these that are more likely to be real, and highPurity is a subset of tight with even stricter requirements. There is a trade-off: loose tracks have high efficiency but also high backgrounds, highPurity has slightly lower efficiency but much lower backgrounds, and tight is in between (see also the plots below). As of CMSSW 7.4, these are all calculated using MVAs (MultiVariate Analysis techniques) for the various iterations. In addition to the status bits, it’s also possible to access the MVA values directly.
Create a new print.py (with a unique name like printMVA.py) file with the following lines:
Add a Handle to the MVA values:
import DataFormats.FWLite as fwlite
events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
tracks = fwlite.Handle("std::vector<reco::Track>")
MVAs = fwlite.Handle("std::vector<float>")
The event loop should be updated to this:
for i, event in enumerate(events):
if i >= 5: break # only the first 5 events
print( "Event", i)
event.getByLabel("generalTracks", tracks)
event.getByLabel("generalTracks", "MVAValues", MVAs)
numTotal = tracks.product().size()
if numTotal == 0: continue
numLoose = 0
numTight = 0
numHighPurity = 0
for j, (track, mva) in enumerate(zip(tracks.product(), MVAs.product())):
if track.quality(track.qualityByName("loose")): numLoose += 1
if track.quality(track.qualityByName("tight")): numTight += 1
if track.quality(track.qualityByName("highPurity")): numHighPurity += 1
if j<50 or 55<j: continue
print( " Track", j, end = " ")
print( "\t charge/pT: %.3f" %(track.charge()/track.pt()), end = " ")
print( "\t phi: %.3f" %track.phi(), end = " ")
print( "\t eta: %.3f" %track.eta(), end = " ")
print( "\t dxy: %.4f" %track.dxy(), end = " ")
print( "\t dz: %.4f" %track.dz(), end = " ")
print( "\t nHits: %s" %track.numberOfValidHits(), "(%s P+ %s S)"%(track.hitPattern().numberOfValidPixelHits(),track.hitPattern().numberOfValidStripHits()), end = " ")
print( "\t algo: %s" %track.algoName(), end = " ")
print( "\t mva: %.3f" %mva)
print( "Event", i, end = " ")
print( "numTotal:", numTotal, end = " ")
print( "numLoose:", numLoose, "(%.1f %%)"%(float(numLoose)/numTotal*100), end = " ")
print( "numTight:", numTight, "(%.1f %%)"%(float(numTight)/numTotal*100), end = " ")
print( "numHighPurity:", numHighPurity, "(%.1f %%)"%(float(numHighPurity)/numTotal*100))
print("total events: ", dir(events))
print("total events: ", type(events))
print("total events: ", events.size())
to print all tracks comment out this line: if j<50 or 55<j: continue as # if j<50 or 55<j: continue
The C++-equivalent is hidden below.
C++ version
Following up in the previous C++ example, the following lines can be added to the PrintOutTracks plugin (the
EDAnalyzer).To class declaration:
edm::EDGetTokenT<edm::View<float> > mvaValsToken_;To constructor:
// after tracksToken_(...) , mvaValsToken_( consumes<edm::View<float> >(iConfig.getUntrackedParameter<edm::InputTag>("mvaValues", edm::InputTag("generalTracks", "MVAValues")) ) )Change your
analyzemethod to:printf("Event %i\n", indexEvent_); edm::Handle<edm::View<reco::Track> > trackHandle; iEvent.getByToken(tracksToken_, trackHandle); if ( !trackHandle.isValid() ) return; const auto numTotal = trackHandle->size(); if (numTotal==0U){ indexEvent_++; return;} edm::Handle<edm::View<float> > trackMVAstoreHandle; iEvent.getByToken(mvaValsToken_,trackMVAstoreHandle); if ( !trackMVAstoreHandle.isValid() ) return; auto numLoose = 0; auto numTight = 0; auto numHighPurity = 0; const edm::View<reco::Track>& tracks = *trackHandle; size_t iTrack = 0; for ( auto track : tracks ) { if (track.quality(track.qualityByName("loose")) ) ++numLoose; if (track.quality(track.qualityByName("tight")) ) ++numTight; if (track.quality(track.qualityByName("highPurity"))) ++numHighPurity; if(iTrack<50 || 55<iTrack){ iTrack++; continue;} printf( "\t Track %zu", iTrack); printf( "\t charge/pT: %.3f", (track.charge()/track.pt())); printf( "\t phi: %.3f", track.phi()); printf( "\t eta: %.3f", track.eta()); printf( "\t dxy: %.4f", track.dxy()); printf( "\t dz: %.4f" , track.dz()); printf( "\t nHits: %i", track.numberOfValidHits()); printf( "(%i P+ %i S)", track.hitPattern().numberOfValidPixelHits(),track.hitPattern().numberOfValidStripHits()); printf( "\t algo: %s" , track.algoName().c_str()); printf( "\t mva: %.3f\n", trackMVAstoreHandle->at(iTrack)); iTrack++; } printf ( "Event %i ", indexEvent_); printf ( "numTotal: %i ", numTotal); printf ( "numLoose: %i ", numLoose); printf ( "(%.1f %%) ",(float(numLoose)/numTotal*100)); printf ( "numTight: %i ", numTight); printf ( "(%.1f %%) ",(float(numTight)/numTotal*100)); printf ( "numHighPurity: %i ", numHighPurity); printf ( "(%.1f %%)\n",(float(numHighPurity)/numTotal*100)); ++indexEvent_;Go back to
TrackingShortExercize/and edit the CMSSW configuration file namedrun_cfg.py(emacs -nw run_cfg.py), adding the following line:process.PrintOutTracks.mvaValues = cms.untracked.InputTag("generalTracks","MVAValues")Now run the
analyzer:cmsRun run_cfg.pyThe
plugincan be found in/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/PrintOutTracks_MVA.ccTheCMSSW configfile can be found in/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run_cfg_MVA.py
Question 1
Now prepare plots for the track variables discussed above, as in the example below (name this file
plot_track_quantities.pyand put it inTrackingShortExercize/). Compare the distributions of track-quality-related variables (number of pixel hits, track goodness of fit, …, which are given in input to MVA classifiers) between tracks passing thehighPurityandLoosequality flags. Do these distributions make sense to you?
Answer
import DataFormats.FWLite as fwlite import ROOT from ROOT import gROOT import os events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root") tracks = fwlite.Handle("std::vector<reco::Track>") hist_pt = ROOT.TH1F("pt", "track pt; p_{T} [GeV]", 100, 0.0, 100.0) hist_eta = ROOT.TH1F("eta", "track eta; #eta", 60, -3.0, 3.0) hist_phi = ROOT.TH1F("phi", "track phi; #phi", 64, -3.2, 3.2) hist_loose_normChi2 = ROOT.TH1F("hist_loose_normChi2" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_loose_numPixelHits = ROOT.TH1F("hist_loose_numPixelHits", "pixel hits; # pixel hits" , 15, 0.0, 15.0) hist_loose_numValidHits = ROOT.TH1F("hist_loose_numValidHits", "valid hits; # valid hits" , 35, 0.0, 35.0) hist_loose_numTkLayers = ROOT.TH1F("hist_loose_numTkLayers" , "valid layers; # valid Tk layers" , 25, 0.0, 25.0) hist_highP_normChi2 = ROOT.TH1F("hist_highP_normChi2" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_highP_numPixelHits = ROOT.TH1F("hist_highP_numPixelHits", "pixel hits; # pixel hits" , 15, 0.0, 15.0) hist_highP_numValidHits = ROOT.TH1F("hist_highP_numValidHits", "valid hits; # valid hits" , 35, 0.0, 35.0) hist_highP_numTkLayers = ROOT.TH1F("hist_highP_numTkLayers" , "valid layers; # valid Tk layers" , 25, 0.0, 25.0) hist_highP_normChi2.SetLineColor(ROOT.kRed) hist_highP_numPixelHits.SetLineColor(ROOT.kRed) hist_highP_numValidHits.SetLineColor(ROOT.kRed) hist_highP_numTkLayers.SetLineColor(ROOT.kRed) for i, event in enumerate(events): event.getByLabel("generalTracks", tracks) for track in tracks.product(): hist_pt.Fill(track.pt()) hist_eta.Fill(track.eta()) hist_phi.Fill(track.phi()) if track.quality(track.qualityByName("loose")): hist_loose_normChi2 .Fill(track.normalizedChi2()) hist_loose_numPixelHits.Fill(track.hitPattern().numberOfValidPixelHits()) hist_loose_numValidHits.Fill(track.hitPattern().numberOfValidHits()) hist_loose_numTkLayers .Fill(track.hitPattern().trackerLayersWithMeasurement()) if track.quality(track.qualityByName("highPurity")): hist_highP_normChi2 .Fill(track.normalizedChi2()) hist_highP_numPixelHits.Fill(track.hitPattern().numberOfValidPixelHits()) hist_highP_numValidHits.Fill(track.hitPattern().numberOfValidHits()) hist_highP_numTkLayers .Fill(track.hitPattern().trackerLayersWithMeasurement()) if i > 500: break gROOT.SetBatch(True) # this allows pyroot to run in batch mode - which prevents the histograms from being displayed every time they are drawn. c = ROOT.TCanvas( "c", "c", 800, 800) # make an output directory odir = "{0}/{1}/".format("plots", "highP") if not os.path.isdir(odir): os.makedirs(odir) # draw and save histograms as pdf files (can alternatively save as png by replacing .pdf with .png hist_pt.Draw() c.SetLogy() c.SaveAs(odir+"track_pt.pdf") c.SetLogy(False) hist_eta.Draw() c.SaveAs(odir+"track_eta.pdf") hist_phi.Draw() c.SaveAs(odir+"track_phi.pdf") hist_highP_normChi2.DrawNormalized() hist_loose_normChi2.DrawNormalized('same') c.SaveAs(odir+"track_normChi2.pdf") hist_highP_numPixelHits.DrawNormalized() hist_loose_numPixelHits.DrawNormalized('same') c.SaveAs(odir+"track_nPixelHits.pdf") hist_highP_numValidHits.DrawNormalized() hist_loose_numValidHits.DrawNormalized('same') c.SaveAs(odir+"track_nValHits.pdf") hist_highP_numTkLayers.DrawNormalized() hist_loose_numTkLayers.DrawNormalized('same') c.SaveAs(odir+"track_nTkLayers.pdf")Optional: save histograms to root file
Additional lines of code to create a root file and save the histograms there
this way you can dynamically view and style histograms (and save to pdf/png with another python script) without having to loop over the AOD/miniAOD everytime you want to change histogram or canvas attributes (like color, line size, labels etc.). Plots are most useful when they clearly describe the information being presented. This technique is also useful for creating “ratio” plots and “pull” plots.
# OPTIONAL: create a root file and save the histograms there ofile = ROOT.TFile.Open(odir+"hists.root", "RECREATE") ofile.WriteObject(hist_pt, "hist_pt") ofile.WriteObject(hist_eta, "hist_eta") ofile.WriteObject(hist_phi, "hist_phi") ofile.WriteObject(hist_loose_normChi2, "hist_loose_normChi2") ofile.WriteObject(hist_highP_normChi2, "hist_highP_normChi2") ofile.WriteObject(hist_loose_numPixelHits, "hist_loose_numPixelHits") ofile.WriteObject(hist_highP_numPixelHits, "hist_highP_numPixelHits") ofile.WriteObject(hist_loose_numValidHits, "hist_loose_numValidHits") ofile.WriteObject(hist_highP_numValidHits, "hist_highP_numValidHits") ofile.WriteObject(hist_loose_numTkLayers, "hist_loose_numTkLayers") ofile.WriteObject(hist_highP_numTkLayers, "hist_highP_numTkLayers")Optional: If having trouble viewing the histograms
You may try displaying the files directly from cmslpc or lxplus using the
displaycommand:display plots/highP/track_pt.pdfthis requires all display settings to the remote connection to be correctly configured.
Sometimes it can be slow to view histograms on cmslpc or lxplus. This can happen because of a slow network connection to the remote computers - possibly when connected far away from Fermilab or CERN.
One option to view histograms is to copy them to the local machine you are working from (i.e. laptop) - this can be done with commands like
scporrsync. You may find more information on these commands by looking at the manuals. From the command line runman scporman rsync. You can also google these command to find more documentation and examples.The basic syntax of both commands (for copying a file from a remote machine to the local machine) is:
<scp/rsync> <options/flags> <source-path> <destination-path>where you must replace the text for each
< >with the relavent information.Recommending to copy files with scp for now (it is faster to learn): First make a directory on the your local computer with
mkdir histograms. And relocate to that dirctory withcd histogramsmkdir histograms cd histograms
- Determine the absolute path on cmslpc of the directory (or file) you wish to have on the local computer:
pwdon cmslpc.- use your cmslpc/lxplus username (if it is the same as on the local machine then ${USER} will work)
- if you set up an ssh config file on the local machine - you can use the hostname from that configuration
The following examples may help guide you to the right command (replace items surrounded by
< >):scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<cmslpc-ssh-hostname>:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:/uscms/homes/<first-letter-of-lpc-username>/<lpc-username>/nobackup/CMSSW_10_6_30_patch1_cmsdas/src/TrackingShortExercize/plots/highP" ./ scp -rp "$USER@cmslpc-el8.fnal.gov:/uscms/homes/${USER:0:1}/${USER}/nobackup/CMSSW_10_6_30_patch1_cmsdas/src/TrackingShortExercize/plots/highP" ./When you have time
rsyncis worth learning about. Note: a main difference betweenscpandrsyncis:scpwill copy all items from the remote machine andrsyncwill only copy the files that don’t exist locally and those that have been updated. Thersynccommand can be very useful - because it can be re-run multiple times without wasting time copying things that are already up to date. The caveat is that you must pay attention to the/at the the end of the path names if you are copying directories for both the . With the/means contents of directory, without the/is including the directory name. This must be considered for both the source path and the destination path.
Track information in MiniAOD
There is no track collection stored in MiniAOD analogous to the generalTracks in AOD. Tracks associated to charged Particle Flow CandidatesPFCandidates are accessible directly from the packedPFCandidates collection for tracks with pT > 0.5 GeV. For tracks between 0.5 and 0.95 GeV the track information is stored with reduced precision. Tracks not associated with PF candidates are in the lostTracks collection if their pT is above 0.95 GeV or they are associated with a secondary vertex or a KS or Lambda candidate. However, for both the tracks associated to the PFCandidates and the lostTracks, the highQuality track selection is used. Tracks with lower quality are not available in MiniAOD at all. In addition, tracks in the lostTracks collection are required to have at least 8 hits of which at least one has to be a pixel hit.
In particular, the track information saved in the PFCandidates is the following:
Track information in PFCandidates collection
- the uncertainty on the impact parameter
dzError(),dxyError()- the number of layers with hits on the track
- the sub/det and layer of first hit of the track
- the
reco::Trackof the candidate is provided by thepseudoTrack()method, with the following information stored:
- the pT,eta and phi of the original track (if those are different from the one of the original
PFCandidate)- an approximate covariance matrix of the track state at the vertex
- approximate
hitPattern()andtrackerExpectedHitsInner()that yield the correct number of hits, pixel hits, layers and the information returned bylostInnerHits()- the track normalized chisquare (truncated to an integer)
- the
highPurityquality flag set, if the original track had it.
Consider that the packedPFCandidates collects both charged and neutral candidates, therefore before trying to access the track information it is important to ensure that the candidate is charged and has the track information correctly stored (track.hasTrackDetails()).
Question 2
Write a simple script
print-comparison.pythat reads a MiniAOD file and the AOD file and compare plots of the same variables we looked at before forHighPuritytracks. For the track pT distributuon, focus on the low pT region below 5 GeV. Can you see any (non-statistical) difference with the previosu plots? The MiniAOD file is located here:root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_MINIAOD.root
Answer
import DataFormats.FWLite as fwlite import ROOT import os ROOT.gROOT.SetBatch(True) # this allows pyroot to run in batch mode - which prevents the histograms from being displayed every time they are drawn. events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_MINIAOD.root") eventsAOD = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root") tracks = fwlite.Handle("std::vector<pat::PackedCandidate>") losttracks = fwlite.Handle("std::vector<pat::PackedCandidate>") tracksAOD = fwlite.Handle("std::vector<reco::Track>") hist_pt = ROOT.TH1F("pt", "track pt; p_{T} [GeV]", 100, 0.0, 100.0) hist_lowPt = ROOT.TH1F("lowPt", "track pt; p_{T} [GeV]", 100, 0.0, 5.0) hist_eta = ROOT.TH1F("eta", "track eta; #eta", 60, -3.0, 3.0) hist_phi = ROOT.TH1F("phi", "track phi; #phi", 64, -3.2, 3.2) hist_normChi2 = ROOT.TH1F("hist_normChi2" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_numPixelHits = ROOT.TH1F("hist_numPixelHits", "pixel hits; # pixel hits" , 15, -0.5, 14.5) hist_numValidHits = ROOT.TH1F("hist_numValidHits", "valid hits; # valid hits" , 35, -0.5, 34.5) hist_numTkLayers = ROOT.TH1F("hist_numTkLayers" , "valid layers; # valid Tk layers" , 25, -0.5, 24.5) hist_pt_AOD = ROOT.TH1F("ptAOD", "track pt; p_{T} [GeV]", 100, 0.0, 100.0) hist_lowPt_AOD = ROOT.TH1F("lowPtAOD", "track pt; p_{T} [GeV]", 100, 0.0, 5.0) hist_eta_AOD = ROOT.TH1F("etaAOD", "track eta; #eta", 60, -3.0, 3.0) hist_phi_AOD = ROOT.TH1F("phiAOD", "track phi; #phi", 64, -3.2, 3.2) hist_normChi2_AOD = ROOT.TH1F("hist_normChi2AOD" , "norm. chi2; norm #chi^{2}" , 100, 0.0, 10.0) hist_numPixelHits_AOD = ROOT.TH1F("hist_numPixelHitsAOD", "pixel hits; # pixel hits" , 15, -0.5, 14.5) hist_numValidHits_AOD = ROOT.TH1F("hist_numValidHitsAOD", "valid hits; # valid hits" , 35, -0.5, 34.5) hist_numTkLayers_AOD = ROOT.TH1F("hist_numTkLayersAOD" , "valid layers; # valid Tk layers" , 25, -0.5, 24.5) hist_pt_AOD.SetLineColor(ROOT.kRed) hist_lowPt_AOD.SetLineColor(ROOT.kRed) hist_eta_AOD.SetLineColor(ROOT.kRed) hist_phi_AOD.SetLineColor(ROOT.kRed) hist_normChi2_AOD.SetLineColor(ROOT.kRed) hist_numPixelHits_AOD.SetLineColor(ROOT.kRed) hist_numValidHits_AOD.SetLineColor(ROOT.kRed) hist_numTkLayers_AOD.SetLineColor(ROOT.kRed) for i, event in enumerate(events): event.getByLabel("packedPFCandidates", "", tracks) event.getByLabel("lostTracks", "", losttracks) alltracks = [track for track in tracks.product()] alltracks += [track for track in losttracks.product()] for track in alltracks : if (not track.hasTrackDetails() or track.charge() == 0 ): continue if not track.trackHighPurity(): continue hist_pt.Fill(track.pt()) hist_lowPt.Fill(track.pt()) hist_eta.Fill(track.eta()) hist_phi.Fill(track.phi()) hist_normChi2 .Fill(track.pseudoTrack().normalizedChi2()) hist_numPixelHits.Fill(track.numberOfPixelHits()) hist_numValidHits.Fill(track.pseudoTrack().hitPattern().numberOfValidHits()) hist_numTkLayers .Fill(track.pseudoTrack().hitPattern().trackerLayersWithMeasurement()) if i > 1000: break for i, event in enumerate(eventsAOD): event.getByLabel("generalTracks", tracksAOD) for j, track in enumerate(tracksAOD.product()) : if not track.quality(track.qualityByName("highPurity")): continue hist_pt_AOD.Fill(track.pt()) hist_lowPt_AOD.Fill(track.pt()) hist_eta_AOD.Fill(track.eta()) hist_phi_AOD.Fill(track.phi()) hist_normChi2_AOD .Fill(track.normalizedChi2()) hist_numPixelHits_AOD.Fill(track.hitPattern().numberOfValidPixelHits()) hist_numValidHits_AOD.Fill(track.hitPattern().numberOfValidHits()) hist_numTkLayers_AOD .Fill(track.hitPattern().trackerLayersWithMeasurement()) if i > 1000: break c = ROOT.TCanvas( "c", "c", 800, 800) # make an output directory odir = "{0}/{1}/".format("plots", "miniAOD") if not os.path.isdir(odir): os.makedirs(odir) # draw and save histograms as pdf files (can alternatively save as png by replacing .pdf with .png hist_pt.Draw() hist_pt_AOD.Draw("same") c.SetLogy() c.SaveAs(odir+"track_pt_miniaod.pdf") hist_lowPt_AOD.Draw() hist_lowPt.Draw("same") c.SetLogy() c.SaveAs(odir+"track_lowPt_miniaod.pdf") c.SetLogy(False) hist_eta_AOD.Draw() hist_eta.Draw("same") c.SaveAs(odir+"track_eta_miniaod.pdf") hist_phi_AOD.Draw() hist_phi.Draw("same") c.SaveAs(odir+"track_phi_miniaod.pdf") hist_normChi2_AOD.Draw() hist_normChi2.Draw("same") c.SaveAs(odir+"track_normChi2_miniaod.pdf") hist_numPixelHits_AOD.Draw() hist_numPixelHits.Draw("same") c.SaveAs(odir+"track_nPixelHits_miniaod.pdf") hist_numValidHits_AOD.Draw() hist_numValidHits.Draw("same") c.SaveAs(odir+"track_nValHits_miniaod.pdf") hist_numTkLayers_AOD.Draw() hist_numTkLayers.Draw("same") c.SaveAs(odir+"track_nTkLayers_miniaod.pdf")Optional: save histograms to root file
see previus example for why you might want to do this.
# OPTIONAL: create a root file and save the histograms there ofile = ROOT.TFile.Open(odir+"hists_AOD.root", "RECREATE") ofile.WriteObject(hist_pt, "hist_pt") ofile.WriteObject(hist_pt_AOD, "hist_pt_AOD") ofile.WriteObject(hist_lowPt, "hist_lowPt") ofile.WriteObject(hist_lowPt_AOD, "hist_lowPt_AOD") ofile.WriteObject(hist_eta, "hist_eta") ofile.WriteObject(hist_eta_AOD, "hist_eta_AOD") ofile.WriteObject(hist_phi, "hist_phi") ofile.WriteObject(hist_phi_AOD, "hist_phi_AOD") ofile.WriteObject(hist_normChi2, "hist_normChi2") ofile.WriteObject(hist_normChi2_AOD, "hist_normChi2_AOD") ofile.WriteObject(hist_numPixelHits, "hist_numPixelHits") ofile.WriteObject(hist_numPixelHits_AOD, "hist_numPixelHits_AOD") ofile.WriteObject(hist_numValidHits, "hist_numValidHits") ofile.WriteObject(hist_numValidHits_AOD, "hist_numValidHits_AOD") ofile.WriteObject(hist_numTkLayers, "hist_numTkLayers") ofile.WriteObject(hist_numTkLayers_AOD, "hist_numTkLayers_AOD")Optional: copy histograms to local computer
Recommending to copy files with scp for now (it is faster to learn): First make a directory on the your local computer with
mkdir histograms. And relocate to that dirctory withcd histogramsmkdir histograms cd histograms
- Determine the absolute path on cmslpc of the directory (or file) you wish to have on the local computer:
pwdon cmslpc.- use your cmslpc/lxplus username (if it is the same as on the local machine then ${USER} will work)
- if you set up an ssh config file on the local machine - you can use the hostname from that configuration
The following examples may help guide you to the right command (replace items surrounded by
< >):scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<cmslpc-ssh-hostname>:<absolute-path-to-cmslpc-directory>" ./ scp -rp "<lpc-username>@cmslpc-el8.fnal.gov:/uscms/homes/<first-letter-of-lpc-username>/<lpc-username>/nobackup/CMSSW_14_0_19/src/TrackingShortExercize/plots/highP" ./ scp -rp "$USER@cmslpc-el8.fnal.gov:/uscms/homes/${USER:0:1}/${USER}/nobackup/CMSSW_14_0_19/src/TrackingShortExercize/plots/highP" ./When you have time
rsyncis worth learning about.
Key Points
The pre-selection of tracks to
HighPurityin MiniAOD also affects the distribution of the track quality parameters.All tracks are stored in the
generalTrackscollection in AOD.In MiniAOD they are accessible in a less straightforward way (
packedPFCandidates,lostTrackscollection) and not all tracks are available!
CMS Data Analysis School Tracking and Vertexing Short Exercise - Tracks as particles
Overview
Teaching: 10 min
Exercises: 10 minQuestions
Can we consider a track as a particle?
Can I use an alternative to the muon object?
Can I define the invariant mass of two tracks?
Objectives
Being familiar with the tracks, particle and identification concepts.
Plot distributions of variables related to a muon resonance.
Unlike calorimeter showers, tracks can usually be interpreted as particle 4-vectors without any additional corrections. Detector alignment, non-helical trajectories from energy loss, Lorentz angle corrections, and (to a much smaller extent) magnetic field inhomogeneities are important, but they are all corrections that must be applied during or before the track-reconstruction process. From an analyzer’s point of view, most tracks are individual particles (depending on quality cuts) and the origin and momentum of the particle are derived from the track’s geometry, with some resolution (random error). Biases (systematic offsets from the true values) are not normal: they’re an indication that something went wrong in this process.
The analyzer does not even need to calculate the particle’s momentum from the track parameters: there are member functions for that. Particle’s transverse momentum, momentum magnitude, and all of its components can be read through the following lines (let’s name this new file kinematics.py and create it in TrackingShortExercize/):
import DataFormats.FWLite as fwlite
import ROOT
import math
events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
tracks = fwlite.Handle("std::vector<reco::Track>")
for i, event in enumerate(events):
event.getByLabel("generalTracks", tracks)
for track in tracks.product():
print( track.pt(), track.p(), track.px(), track.py(), track.pz())
if i > 20: break
Question 1
Now we can investigate the kinematics of the tracks in our file. Assuming that the particle is a pion (pion mass = 0.140 GeV), calculate its kinetic energy.
Additional information
Identifying the particle that made the track is difficult: the mass of some low-momentum tracks can be identified by their energy loss, called dE/dx, and electrons and muons can be identified by signatures in other subdetectors. Without any other information, as an approximation, we can assume that a randomly chosen track is a pion, since hadron collisions produce a lot of pions.
Answer
import DataFormats.FWLite as fwlite import ROOT import math events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root") tracks = fwlite.Handle("std::vector<reco::Track>") for i, event in enumerate(events): event.getByLabel("generalTracks", tracks) for track in tracks.product(): print( track.pt(), track.p(), track.px(), track.py(), track.pz()) print( "energy: ", math.sqrt(0.140**2 + track.p()**2)) if i > 20: break
Now, let’s look for resonances! Given two tracks,
if len(tracks.product()) > 1:
one = tracks.product()[0]
two = tracks.product()[1]
the invariant mass may be calculated as
total_energy = math.sqrt(0.140**2 + one.p()**2) + math.sqrt(0.140**2 + two.p()**2)
total_px = one.px() + two.px()
total_py = one.py() + two.py()
total_pz = one.pz() + two.pz()
mass = math.sqrt(total_energy**2 - total_px**2 - total_py**2 - total_pz**2)
However, this quantity has no meaning unless the two particles are actually descendants of the same decay. Two randomly chosen tracks (out of hundreds per event) typically are not.
To increase the chances that pairs of randomly chosen tracks are descendants of the same decay, consider a smaller set of tracks: muons. Muons are identified by the fact that they can pass through meters of iron (the CMS magnet return yoke), so muon tracks extend from the silicon tracker to the muon chambers (see CMS quarter-view below), as much as 12 meters long! Muons are rare in hadron collisions. If an event contains two muons, they often (though not always) come from the same decay.
![]()
Normally, one would access muons through the reco::Muon object since this contains additional information about the quality of the muon hypothesis. For simplicity, we will access their track collection in the same way that we have been accessing the main track collection. We only need to replace generalTracks with globalMuons. Add the following loop to kinematics.py.
events.toBegin()
for i, event in enumerate(events):
if i >= 15: break # only the first 15 events
print( "Event", i)
event.getByLabel("globalMuons", tracks)
for j, track in enumerate(tracks.product()):
print( " Track", j, track.charge()/track.pt(), track.phi(), track.eta(), track.dxy(), track.dz())
Run this code on the run321167_Charmonium_AOD.root file found here:
root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_Charmonium_AOD.root
Notice how few muon tracks there are compared to the same code executed for generalTracks. In fact, you only see as many muons as you do because this data sample was collected with a muon trigger. (The muon definition in the trigger is looser than the globalMuons algorithm, which is why there are some events with fewer than two globalMuons.)
See in the Appendix an application for the Muon and Tracks objects usage in the CMS tracking efficiency computation.
Question 2
As an exercise, make a histogram of all di-muon masses from 0 to 5 GeV. Exclude events that do not have exactly two muon tracks, and note that the muon mass is 0.106 GeV). Create a file
dimuon_mass.pyinTrackingShortExercize/for this purpose.
More…
The solution combines several of the techniques introduced above:
import math import DataFormats.FWLite as fwlite import ROOT import os ROOT.gROOT.SetBatch(True) events = fwlite.Events("root://cmseos.fnal.gov//store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_Charmonium_AOD.root") tracks = fwlite.Handle("std::vector<reco::Track>") mass_histogram = ROOT.TH1F("mass", "mass", 100, 0.0, 5.0) events.toBegin() for event in events: event.getByLabel("globalMuons", tracks) product = tracks.product() if product.size() == 2: one = product[0] two = product[1] if not (one.charge()*two.charge() == -1): continue energy = (math.sqrt(0.106**2 + one.p()**2) + math.sqrt(0.106**2 + two.p()**2)) px = one.px() + two.px() py = one.py() + two.py() pz = one.pz() + two.pz() mass = math.sqrt(energy**2 - px**2 - py**2 - pz**2) mass_histogram.Fill(mass) c = ROOT.TCanvas ("c", "c", 800, 800) odir = "{0}/{1}/".format("plots", "dimuonmass") if not os.path.isdir(odir): os.makedirs(odir) mass_histogram.Draw() c.SaveAs(odir+"mass.png")The histogram should look like this:
Key Points
Tracks give us a direct handle on actual particles, thus they can easily be used to reconstruct other particles in the event
CMS Data Analysis School Tracking and Vertexing Short Exercise - Constructing vertices from tracks
Overview
Teaching: 20 min
Exercises: 20 minQuestions
What is a (primary, secondary) vertex object? What about beamspot?
Which information are available in different CMS data formats?
How can I run the vertex reconstruction?
Can we get better physics results using primary verteces?
Objectives
Being familiar with the vertex concept.
Plot distributions about the main vertex variables.
Open the stage for Appendix exercises.
In this exercise, we will encounter three main objects: primary vertices, secondary vertices, and the beamspot. Let’s start with secondary vertices. Particles produced by a single decay or collision radiate from the point of their origin. Their tracks should nearly cross at this point (within measurement uncertainties): if two or more tracks cross at some point, it is very likely that they descend from the same decay or collision.
This test is more significant than it may appear by looking at event pictures. With so many tracks, it looks like they cross accidentally, but the two-dimensional projection of the event picture is misleading. One-dimensional paths through three-dimensional space do not intersect easily, especially when the trajectories of those paths are measured with microns to hundreds-of-microns precision.
Starting from the detected tracks, we work backward and reconstruct the original vertices by checking each pair of tracks for overlaps. This is performed in the standard reconstruction sequence and delivered to the analyst as lists of KS → π+π- and Λ → pπ0 candidates, but we will repeat the procedure with different parameters. The algorithm will run three times, the first accepting all (loose) tracks, the second accepting only tight tracks, and the third accepting only highPurity tracks.
These are called secondary vertices because the proton-proton collision produced the first (primary) vertex, then the neutral KS flew several centimeters away from the rest of the collision products and decayed into π+π- at a second (secondary) position in space.
Running the vertex reconstruction
Create a file named construct_secondary_vertices_cfg.py in TrackingShortExercize/ and fill it with the following:
import FWCore.ParameterSet.Config as cms
process = cms.Process("KSHORTS")
# Use the tracks_and_vertices.root file as input.
process.source = cms.Source("PoolSource",
fileNames = cms.untracked.vstring("file:/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_Charmonium_AOD.root"))
process.maxEvents = cms.untracked.PSet(input = cms.untracked.int32(500))
# Suppress messages that are less important than ERRORs.
process.MessageLogger = cms.Service("MessageLogger",
destinations = cms.untracked.vstring("cout"),
cout = cms.untracked.PSet(threshold = cms.untracked.string("ERROR")))
# Load part of the CMSSW reconstruction sequence to make vertexing possible.
# We'll need the CMS geometry and magnetic field to follow the true, non-helical
# shapes of tracks through the detector.
process.load("Configuration/StandardSequences/FrontierConditions_GlobalTag_cff")
from Configuration.AlCa.GlobalTag import GlobalTag
process.GlobalTag = GlobalTag(process.GlobalTag, "auto:run2_data")
process.load("Configuration.Geometry.GeometryRecoDB_cff")
process.load("Configuration.StandardSequences.MagneticField_cff")
process.load("TrackingTools.TransientTrack.TransientTrackBuilder_cfi")
# Copy most of the vertex producer's parameters, but accept tracks with
# progressively more strict quality.
process.load("RecoVertex.V0Producer.generalV0Candidates_cfi")
# loose
process.SecondaryVerticesFromLooseTracks = process.generalV0Candidates.clone(
trackRecoAlgorithm = cms.InputTag("generalTracks"),
doKshorts = cms.bool(True),
doLambdas = cms.bool(True),
trackQualities = cms.string("loose"),
innerHitPosCut = cms.double(-1.),
cosThetaXYCut = cms.double(-1.),
)
# tight
process.SecondaryVerticesFromTightTracks = process.SecondaryVerticesFromLooseTracks.clone(
trackQualities = cms.string("tight"),
)
# highPurity
process.SecondaryVerticesFromHighPurityTracks = process.SecondaryVerticesFromLooseTracks.clone(
trackQualities = cms.string("highPurity"),
)
# Run all three versions of the algorithm.
process.path = cms.Path(process.SecondaryVerticesFromLooseTracks *
process.SecondaryVerticesFromTightTracks *
process.SecondaryVerticesFromHighPurityTracks)
# Writer to a new file called output.root. Save only the new K-shorts and the
# primary vertices (for later exercises).
process.output = cms.OutputModule(
"PoolOutputModule",
SelectEvents = cms.untracked.PSet(SelectEvents = cms.vstring("path")),
outputCommands = cms.untracked.vstring(
"drop *",
"keep *_*_*_KSHORTS",
"keep *_offlineBeamSpot_*_*",
"keep *_offlinePrimaryVertices_*_*",
"keep *_offlinePrimaryVerticesWithBS_*_*",
),
fileName = cms.untracked.string("output.root")
)
process.endpath = cms.EndPath(process.output)
The name of the secondary-vertex producer (process.SecondaryVerticesFromLooseTracks) given in this configuration will be the name of the produced collection with process. stripped off (see the usage below).
Run the configuration file with:
cmsRun construct_secondary_vertices_cfg.py
which can take a couple of minutes to complete. Notice it will create the output.root file which will be used later.
You can check the content of the file by running the simple script as follows
edmDumpEventContent output.root
Looking at secondary vertices
When it’s done, you can open output.root and read the vertex positions, just as you did for the track momenta with the following lines (let’s name this code sec_vertices.py and put it in TrackingShortExercize/):
import DataFormats.FWLite as fwlite
events = fwlite.Events("file:output.root")
secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositeCandidate>")
events.toBegin()
for i, event in enumerate(events):
print( "Event:", i)
event.getByLabel("SecondaryVerticesFromLooseTracks", "Kshort", secondaryVertices)
for j, vertex in enumerate(secondaryVertices.product()):
print( " Vertex:", j, vertex.vx(), vertex.vy(), vertex.vz())
if i > 10: break
In the code above, you specify the C++ type of the collection (std::vector). For each event you obtain the collection of secondary vertices originating from KS decays, by providing the name of the collection producer (SecondaryVerticesFromLooseTracks) and the label for the KS collection (Kshort, defined here).
Each of these vertices contains two tracks by construction. One of the vertex member functions (do dir(vertex) in the python shell after executing the above code to see them all) returns the invariant mass of the pair of tracks. This calculation is a little more complex than the invariant masses we calculated by hand because it is necessary to evaluate the px and py components of momentum at the vertex rather than at the beamspot. If these two particles really did originate in a decay several centimeters from the beamspot, then the track parameters evaluated at the beamspot are not meaningful.
Question 1
Plot the invariant mass of all vertices. Modify the
sec_vertices.pyto draw the invariant mass distribution, i.e. fill a histogram withvertex.mass()for each secondary vertex.
Answer (don’t look until you’ve tried it!)
import DataFormats.FWLite as fwlite import ROOT events = fwlite.Events("file:output.root") secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositeCandidate>") mass_histogram = ROOT.TH1F("mass", "mass", 100, 0.4, 0.6) events.toBegin() for event in events: event.getByLabel("SecondaryVerticesFromLooseTracks", "Kshort", secondaryVertices) for vertex in secondaryVertices.product(): mass_histogram.Fill(vertex.mass()) c = ROOT.TCanvas ("c" , "c", 800, 800) mass_histogram.Draw() c.SaveAs("kshort_mass.png")
Question 2
You should see a very prominent KS → π+π- peak, but also a pedestal. What is the pedestal? Why does it cut off at 0.43 and 0.57 GeV?
More
You can answer the question concerning the cut-off with the information here.
Question 3
Prepare a similar plot for the Lambda vertices. (Hints: were the Lambda vertices created when you ran the SecondaryVertices producer? the Lambda mass is 1.115 GeV. Are you expecting to reconstruct the mass at the same value in the whole detector ? (optionally plot the mass resolution as function of the eta of the reconstructed Lambda).
Question 4
Plot the flight distance of all vertices. Modify the
sec_vertices.pyto draw the flight distance in the transverse plane distribution between each secondary vertex and the primary vertex (PV). For this, you can look ahead at the next part of the exercise to see how to access the primary vertices collection. Once you have it, you can access the first primary vertex in the collection withpv = primaryVertices.product()[0]. You can now use the x and y coordinates of the secondary vertices and the primary vertex to calculate the distance. Note that for the PV, you access these values with thex()andy()member functions, while for the secondary vertices, these are calledvx()andvy().
Answer
import DataFormats.FWLite as fwlite import ROOT events = fwlite.Events("file:output.root") secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositeCandidate>") primaryVertices = fwlite.Handle("std::vector<reco::Vertex>") lxy_histogram = ROOT.TH1F("lxy", "lxy", 500, 0, 70) events.toBegin() for event in events: event.getByLabel("offlinePrimaryVertices", primaryVertices) pv = primaryVertices.product()[0] event.getByLabel("SecondaryVerticesFromLooseTracks", "Lambda", secondaryVertices) for vertex in secondaryVertices.product(): lxy = ((vertex.vx()-pv.x())**2 + (vertex.vy() - pv.y())**2)**0.5 lxy_histogram.Fill(lxy) c = ROOT.TCanvas ("c" , "c", 800, 800) lxy_histogram.Draw() c.SaveAs("lambda_lxy.png")
Question 5
You should rerun the
construct_secondary_vertices.pyto process all events in the file to get more statistics. For this, set themaxEventsparameter to-1. Running on more events, one can appreciate some structures in the flight distance distribution, can you explain them ? Note that these are especially noticeable in the distribution of Lambdas.
Secondary vertices in MiniAOD
The SecondaryVertex collection is already available in the MiniAOD files. You can retrieve the collection type and format by doing:
cp /eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_Charmonium_MINIAOD.root $TMPDIR
edmDumpEventContent $TMPDIR/run321167_Charmonium_MINIAOD.root | grep Vertices
Question 6
Modify
sec_vertices.pyto read the lambda vertices from the MiniAOD file. You will have to modify the collection type and label accordingly to the MiniAOD content.
Answer
import DataFormats.FWLite as fwlite import ROOT events = fwlite.Events("/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_Charmonium_MINIAOD.root") secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositePtrCandidate>") mass_histogram = ROOT.TH1F("mass_histogram", "mass_histogram", 100, 1., 1.2) events.toBegin() for i, event in enumerate(events): event.getByLabel("slimmedLambdaVertices", "", secondaryVertices) for j, vertex in enumerate(secondaryVertices.product()): mass_histogram.Fill(vertex.mass()) c = ROOT.TCanvas() mass_histogram.Draw() c.SaveAs("mass_lambda_miniaod.png")
Basic distributions of primary vertices
The primary vertex reconstruction consists of three steps:
Steps of primary vertex reconstruction
- Selection of tracks
- Clustering of the tracks that appear to originate from the same interaction vertex
- Fitting for the position of each vertex using its associated tracks
All the primary vertices reconstructed in an event are saved in the reco::Vertex collection labeled offlinePrimaryVertices. Create the file vertex.py in TrackingShortExercize/ which will load the original run321167_ZeroBias_AOD.root file and make a quarter-view plot of the vertex distribution (run it using python3 vertex.py):
import DataFormats.FWLite as fwlite
import math
import ROOT
events = fwlite.Events("/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
primaryVertices = fwlite.Handle("std::vector<reco::Vertex>")
rho_z_histogram = ROOT.TH2F("rho_z", "rho_z", 100, 0.0, 30.0, 100, 0.0, 10.0)
events.toBegin()
for event in events:
event.getByLabel("offlinePrimaryVertices", primaryVertices)
for vertex in primaryVertices.product():
rho_z_histogram.Fill(abs(vertex.z()),
math.sqrt(vertex.x()**2 + vertex.y()**2))
c = ROOT.TCanvas("c", "c", 800, 800)
rho_z_histogram.Draw()
c.SaveAs("rho_z.png")
You should see a broad distribution in z, close to rho = 0 (much closer than for the secondary vertices, see appendix). In fact, the distribution is about 0.1 cm wide and 4 cm long. The broad distribution in z is helpful: if 20 primary vertices are uniformly distributed along a 4 cm pencil-like region, we only need 2 mm vertex resolution to distinguish neighboring vertices. Fortunately, the CMS vertex resolution is better than this (better than 20μm and 25μm in x and z, respectively TRK-11-001, performances with the Phase1 pixel detector), so they can be distinguished with high significance.
To see this, you should modify properly the previous code to make a plot of the distance between primary vertices:
deltaz_histogram = ROOT.TH1F("deltaz", "deltaz", 1000, -20.0, 20.0)
events.toBegin()
for event in events:
event.getByLabel("offlinePrimaryVertices", primaryVertices)
pv = primaryVertices.product()
for i in xrange(pv.size() - 1):
for j in xrange(i + 1, pv.size()):
deltaz_histogram.Fill(pv[i].z() - pv[j].z())
c = ROOT.TCanvas ("c", "c", 800, 800)
deltaz_histogram.Draw()
c.SaveAs("deltaz.png")
The broad distribution is due to the spread in primary vertex positions. Zoom in on the narrow dip near deltaz = 0. This region is empty because if two real vertices are too close to each other, they will be misreconstructed as a single vertex. Thus, there are no reconstructed vertices with such small separations.
Question 7
Write a short script to print out the number of primary vertices in each event. When people talk about the pile-up, it is this number they are referring to. If you want, you can even plot it; the distribution should roughly fit a Poisson distribution.
Answer
events.toBegin() for i, event in enumerate(events): event.getByLabel("offlinePrimaryVertices", primaryVertices) print( "Pile-up:", primaryVertices.product().size()) if i > 100: break
Print out the number of tracks in a single vertex object. (Use the trick for vertex in primaryVertices.product(): break to obtain a primary vertex object.)
Solution
print( vertex.nTracks()) print( vertex.tracksSize())Why might they be different? ( See tracksSize vs nTracks).
Note:in C++, you could loop over the tracks associated with this vertex, but this functionality doesn’t work in Python. In the standard analysis workflow there are many quality requirements to be applied to the events and to the reconstructed quantities in an event. One of these requests is based on the characteristics of the reconstructed primary vertices, and it is defined by theCMSSW EDFiltergoodOfflinePrimaryVertices_cfi.py . Are these selections surprising you ?
Question 8
Write a short script to plot the distribution of the number of tracks vs the number of vertices. What do you expect?
Answer
The following snippet tells you what to do, but you need to make sure that all needed variables are defined.
histogram = ROOT.TH2F("ntracks_vs_nvertex","ntracks_vs_nvertex", 30, 0.0, 29.0, 100, 0.0,2000.0) events.toBegin() for event in events: event.getByLabel("offlinePrimaryVertices", primaryVertices) event.getByLabel("generalTracks", tracks) histogram.Fill(primaryVertices.product().size(), tracks.product().size()) c = ROOT.TCanvas("c", "c", 800, 800) histogram.Draw() c.SaveAs("ntracks_vs_nvertex.png")
It’s also useful to distinguish between the primary vertices and the beamspot. A read-out event can have many primary vertices, each of which usually corresponds to a point in space where two protons collide. The beamspot is an estimate of where protons are expected to collide, derived from the distribution of primary vertices. Not only does an event have only one beamspot, but the beamspot is constant for a lumi-section (time interval of 23.31 consecutive seconds). This script prints the average x-position of the primary vertices and plots their x-y distribution (name it beamspot.py and put it in TrackingShortExercize):
import DataFormats.FWLite as fwlite
import math
import ROOT
def isGoodPV(vertex):
if ( vertex.isFake() or
vertex.ndof < 4.0 or
abs(vertex.z()) > 24.0 or
abs(vertex.position().Rho()) > 2):
return False
return True
events = fwlite.Events("/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
primaryVertices = fwlite.Handle("std::vector<reco::Vertex>")
beamspot = fwlite.Handle("reco::BeamSpot")
vtx_xy = ROOT.TH2F('vtx_xy','; x [cm]; y [cm]', 100,-0.1, 0.3, 100, -0.25, 0.15)
sumx = 0.0
N = 0
last_beamspot = None
events.toBegin()
for event in events:
event.getByLabel("offlinePrimaryVertices", primaryVertices)
event.getByLabel("offlineBeamSpot", beamspot)
if last_beamspot == None or last_beamspot != beamspot.product().x0():
print( "New beamspot IOV (interval of validity)...")
last_beamspot = beamspot.product().x0()
sumx = 0.0
N = 0
for vertex in primaryVertices.product():
if not isGoodPV(vertex): continue
N += 1
sumx += vertex.x()
vtx_xy.Fill(vertex.x(), vertex.y())
if N % 1000 == 0:
print( "Mean of primary vertices:", sumx/N, end = " ")
print( "Beamspot:", beamspot.product().x0())
c = ROOT.TCanvas( "c", "c", 1200, 800)
vtx_xy.Draw("colz")
c.SaveAs("vtx_xy.png")
Add the analogous 2D plots for x versus z and y vs z positions.
More exercises…
Furthermore, you can add a plot of the average primary vertex position and compare it to:
- the center of beamspot (red line)
- the beamspot region (beamspot center +/- beamspot witdh, that you can access through
beamspot.product().BeamWidthX()
Answer
import DataFormats.FWLite as fwlite import math import ROOT from array import array ROOT.gROOT.SetBatch(True) def isGoodPV(vertex): if ( vertex.isFake() or \ vertex.ndof < 4.0 or \ abs(vertex.z()) > 24.0 or \ abs(vertex.position().Rho()) > 2): return False return True events = fwlite.Events("/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root") primaryVertices = fwlite.Handle("std::vector<reco::Vertex>") beamspot = fwlite.Handle("reco::BeamSpot") vtx_position, N_vtx = array( 'd' ), array( 'd' ) vtx_xy = ROOT.TH2F('vtx_xy','; x [cm]; y [cm]', 100,-0.05, 0.25, 100, -0.2, 0.1) c = ROOT.TCanvas( "c", "c", 1200, 800) sumx = 0.0 N = 0 iIOV = 0 last_beamspot = None last_beamspot_sigma = None vtx_position, N_vtx = array( 'd' ), array( 'd' ) leg = ROOT.TLegend(0.45, 0.15, 0.6, 0.28) leg.SetBorderSize(0) leg.SetTextSize(0.03) events.toBegin() for event in events: event.getByLabel("offlinePrimaryVertices", primaryVertices) event.getByLabel("offlineBeamSpot", beamspot) if last_beamspot == None or last_beamspot != beamspot.product().x0(): print( "New beamspot IOV (interval of validity)...") ## first save tgraph and then reset if (iIOV > 0): theGraph = ROOT.TGraph(len(vtx_position), N_vtx, vtx_position) theGraph.SetMarkerStyle(8) theGraph.SetTitle('IOV %s; N Vtx; X position'%iIOV) theGraph.Draw('AP') theGraph.GetYaxis().SetRangeUser(0.094, 0.100) line = ROOT.TLine(0,last_beamspot,N_vtx[-1],last_beamspot) line.SetLineColor(ROOT.kRed) line.SetLineWidth(2) line.Draw() line2 = ROOT.TLine(0,last_beamspot - last_beamspot_sigma,N_vtx[-1],last_beamspot - last_beamspot_sigma) line2.SetLineColor(ROOT.kOrange) line3 = ROOT.TLine(0,last_beamspot + last_beamspot_sigma,N_vtx[-1],last_beamspot + last_beamspot_sigma) line3.SetLineColor(ROOT.kOrange) line2.SetLineWidth(2) line3.SetLineWidth(2) line2.Draw() line3.Draw() leg.Clear() leg.AddEntry(theGraph, 'ave. vtx position', 'p') leg.AddEntry(line , 'center of the beamspot ' , 'l') leg.AddEntry(line2 , 'center of the bs #pm beamspot width ' , 'l') leg.Draw() c.SaveAs("vtx_x_vs_N_%s.png"%iIOV) break vtx_position, N_vtx = array( 'd' ), array( 'd' ) last_beamspot = beamspot.product().x0() last_beamspot_sigma = beamspot.product().BeamWidthX() sumx = 0.0 N = 0 iIOV += 1 for vertex in primaryVertices.product(): if not isGoodPV(vertex): continue N += 1 sumx += vertex.x() if N % 1000 == 0: vtx_position.append(sumx/N) N_vtx.append(N)
Primary vertices improve physics results
Finally, let’s consider an example of how primary vertices are useful to an analyst. If you’re interested in KS → π+π-, it might seem that primary vertices are irrelevant because neither of your two visible tracks (π+π-) directly originated in any of the proton-proton collisions. However, the KS did. The KS flew several centimeters away from the primary vertex in which it was produced, and its direction of flight must be parallel to its momentum (by definition).
We can measure the KS momentum from the momenta of its decay products, and we can identify the start and end positions of the KS flight from the locations of the primary and secondary vertices. The momentum vector and the displacement vector must be parallel. (There is no constraint on the length of the displacement vector because the lifetimes of KS mesons follow an exponentially random distribution.)
Create a new file analyse.py in TrackingShortExercize/ which will open output.root:
import math
import DataFormats.FWLite as fwlite
import ROOT
events = fwlite.Events("file:output.root")
primaryVertices = fwlite.Handle("std::vector<reco::Vertex>")
secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositeCandidate>")
Create a histogram in which to plot the distribution of cosAngle, which is the normalized dot product of the KS momentum vector and its displacement vector. Actually, make two histograms to zoom into the cosAngle → 1 region.
cosAngle_histogram = ROOT.TH1F("cosAngle", "cosAngle", 100, -1.0, 1.0)
cosAngle_zoom_histogram = ROOT.TH1F("cosAngle_zoom", "cosAngle_zoom", 100, 0.99, 1.0)
Question 9
Construct a nested loop over secondary and primary vertices to compute displacement vectors and compare them with KS momentum vectors. Just print out a few values.
Answer
for i, event in enumerate(events): event.getByLabel("offlinePrimaryVertices", primaryVertices) event.getByLabel("SecondaryVerticesFromLooseTracks", "Kshort", secondaryVertices) for secondary in secondaryVertices.product(): px = secondary.px() py = secondary.py() pz = secondary.pz() p = secondary.p() for primary in primaryVertices.product(): dx = secondary.vx() - primary.x() dy = secondary.vy() - primary.y() dz = secondary.vz() - primary.z() dl = math.sqrt(dx**2 + dy**2 + dz**2) print( "Normalized momentum:", px/p, py/p, pz/p, end = " ") print( "Normalized displacement:", dx/dl, dy/dl, dz/dl) if i > 20: break
For each set of primary vertices, find the best cosAngle and fill the histograms with that.
Solution
import math import DataFormats.FWLite as fwlite import ROOT events = fwlite.Events("file:output.root") primaryVertices = fwlite.Handle("std::vector<reco::Vertex>") secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositeCandidate>") cosAngle_histogram = ROOT.TH1F("cosAngle", "cosAngle", 100, -1.0, 1.0) cosAngle_zoom_histogram = ROOT.TH1F("cosAngle_zoom", "cosAngle_zoom", 100, 0.99, 1.0) events.toBegin() for event in events: event.getByLabel("offlinePrimaryVertices", primaryVertices) event.getByLabel("SecondaryVerticesFromLooseTracks", "Kshort", secondaryVertices) for secondary in secondaryVertices.product(): px = secondary.px() py = secondary.py() pz = secondary.pz() p = secondary.p() bestCosAngle = -1 # start with the worst possible for primary in primaryVertices.product(): dx = secondary.vx() - primary.x() dy = secondary.vy() - primary.y() dz = secondary.vz() - primary.z() dl = math.sqrt(dx**2 + dy**2 + dz**2) dotProduct = px*dx + py*dy + pz*dz cosAngle = dotProduct / p / dl if cosAngle > bestCosAngle: bestCosAngle = cosAngle # update it if you've found a better one cosAngle_histogram.Fill(bestCosAngle) cosAngle_zoom_histogram.Fill(bestCosAngle) c = ROOT.TCanvas("c" , "c" , 800, 800) cosAngle_histogram.Draw() c.SaveAs("cosAngle.png") cosAngle_zoom_histogram.Draw() c.SaveAs("cosAngle_zoom.png")
Finally, create KS mass histograms, with and without requiring bestCosAngle to be greater than 0.99. Does this improve the signal-to-background ratio?
Solution
import math import DataFormats.FWLite as fwlite import ROOT events = fwlite.Events("file:output.root") primaryVertices = fwlite.Handle("std::vector<reco::Vertex>") secondaryVertices = fwlite.Handle("std::vector<reco::VertexCompositeCandidate>") mass_histogram = ROOT.TH1F("mass", "mass", 100, 0.4, 0.6) cosAngle_histogram = ROOT.TH1F("cosAngle", "cosAngle", 100, -1.0, 1.0) cosAngle_zoom_histogram = ROOT.TH1F("cosAngle_zoom", "cosAngle_zoom", 100, 0.99, 1.0) mass_goodCosAngle = ROOT.TH1F("mass_goodCosAngle", "mass_goodCosAngle", 100, 0.4, 0.6) events.toBegin() for event in events: event.getByLabel("offlinePrimaryVertices", primaryVertices) event.getByLabel("SecondaryVerticesFromLooseTracks", "Kshort", secondaryVertices) for secondary in secondaryVertices.product(): px = secondary.px() py = secondary.py() pz = secondary.pz() p = secondary.p() bestCosAngle = -1 # start with the worst possible for primary in primaryVertices.product(): dx = secondary.vx() - primary.x() dy = secondary.vy() - primary.y() dz = secondary.vz() - primary.z() dl = math.sqrt(dx**2 + dy**2 + dz**2) dotProduct = px*dx + py*dy + pz*dz cosAngle = dotProduct / p / dl if cosAngle > bestCosAngle: bestCosAngle = cosAngle # update it if you've found a better one cosAngle_histogram.Fill(bestCosAngle) cosAngle_zoom_histogram.Fill(bestCosAngle) if bestCosAngle > 0.99: mass_goodCosAngle.Fill(secondary.mass()) mass_histogram.Fill(secondary.mass()) c = ROOT.TCanvas("c", "c", 800, 800) mass_histogram.Draw() mass_goodCosAngle.SetLineColor(ROOT.kRed) mass_goodCosAngle.Draw("same") c.SaveAs("mass_improved.png")
That’s all! The session is over, unless you would like to try some of the extra questions and arguments listed in the Appendix!
Key Points
Two or more tracks can be used to reconstruct their common point of origin, the vertex
Requiring two tracks to originate from the same vertex is a powerful tool to identify particles that decayed in the detector
The distribution of the proton proton interaction, the so-called interaction region, in the center of the CMS detector is an ellipse.
CMS Data Analysis School Tracking and Vertexing Short Exercise - Appendix
Overview
Teaching: 0 min
Exercises: 1 minQuestions
How can I use track variables to retrieve the CMS tracking efficiency?
Can I find more about secondary vertex?
Is there a correlation between pile-up and number of clusters?
Objectives
After the in-person session, being familiar with more information about tracks, vertex and CMS tracking detector.
LXR: a tool to search through CMSSW code
LXR is a very useful tool to look-up methods/classes/configuration files/parameters/example etc. when working on software development in CMS.
- CERN: https://cmssdt.cern.ch/lxr/
- Fermilab: http://cmslxr.fnal.gov/lxr/
Further useful code references
- CMSSW source code on GitHub for
CMSSW_14_0_19: https://github.com/cms-sw/cmssw/tree/CMSSW_14_0_19- you can switch to the branch or version (encoded as git tag) using the drop down menu left of the green
New Pull Requestbutton
- you can switch to the branch or version (encoded as git tag) using the drop down menu left of the green
- CMSSW Reference Manual: https://cmsdoxygen.web.cern.ch/cmsdoxygen/CMSSW_14_0_19/doc/html/classes.html
Tracking efficiency performance via the Tag and Probe technique
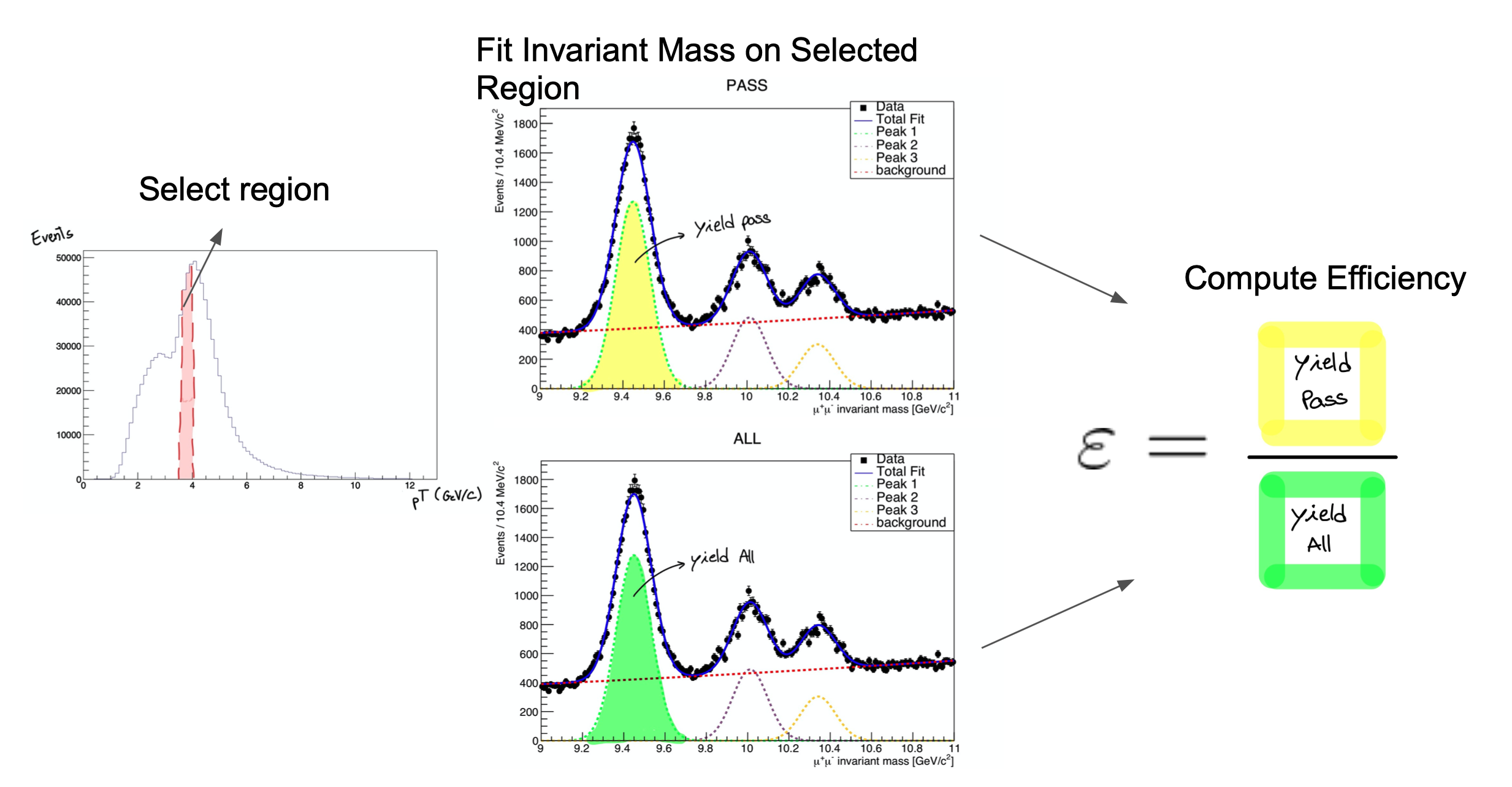
The tag and probe method is a data-driven technique for measuring particle detection efficiencies. This approach is broadly used in obtaining trigger, tracking, identification and many more steps efficiency. It is based on the decays of known resonances (e.g. J/ψ, ϒ and Z) to pairs of the particles being studied. In this exercise, these particles are muons, and the Z resonance is nominally used. The determination of the detector efficiency is a critical ingredient in any physics measurement. It accounts for the particles that were produced in the collision but escaped detection (did not reach the detector elements, were missed by the reconstructions algorithms, etc). It can be in general estimated using simulations, but simulations need to be calibrated with data. The T&P method here described provides a useful and elegant mechanism for extracting efficiencies directly from data!
What is “tag” and “probe”?
The resonance, used to calculate the efficiencies, decays to a pair of particles: the tag and the probe.
- Tag muon = well identified, triggered muon (tight selection criteria).
- Probe muon = unbiased set of muon candidates (very loose selection criteria), either passing or failing the criteria for which the efficiency is to be measured.
How do we calculate the efficiency?
The efficiency is given by the fraction of probe muons that pass a given criteria:
- The denominator corresponds to the number of resonance candidates (tag+probe pairs) reconstructed in the dataset.
- The numerator corresponds to the subset for which the probe passes the criteria.
- The tag+probe invariant mass distribution is used to select only signal, that is, only true Z candidates decaying to dimuons. This is achieved in this exercise by the usage of the fitting method.
In this exercise the probe muons are StandAlone muons: all tracks of the segments reconstructed in the muon chambers (performed using segments and hits from Drift Tubes - DTs in the barrel region, Cathode strip chambers - CSCs in the endcaps and Resistive Plates Chambers - RPCs for all muon system) are used to generate “seeds” consisting of position and direction vectors and an estimate of the muon transverse momentum. The standalone muon is matched in (ΔR < 0.3, Δη < 0.3) with generalTracks in AOD, lostTracks in MiniAOD having pT larger than 10 GeV, being in this case a passing probes.
The fitting method
After seelcting the tag & probe pair events, the data can be put into 2 categories: probe muons passing seelction, and all probes. In both of the 2 categories, to better etimate the true signal contribution, we do a fit on the a kinematic variable instead of simply counting the number of events.
The procedure is applied after splitting the data in bins of a kinematic variable of the probe object (e.g. the traverse momentum, pT); as such, the efficiency will be measured as a function of that quantity for each of the bins. Usually we obtain different efficiency in different pT or η bins in tracking efficiency measurement, because different pT and η particles curve differently and tend to have different detector behaviours. So, in the picture below, on the left, let’s imagine that the pT bin we are selecting is the one marked in red. But, of course, in that bin (like in the rest) you will have true Z decays as well as muon pairs from other processes (maybe QCD, for instance). The true decays would make up our signal, whereas the other events will be considered the background.
The fit, which is made in a different space (the invariant mass space) allows to statistically discriminate between signal and background. To compute the efficiency we simply divide the signal yield from the fits to the passing category by the signal yield from the fit of the inclusive (All) category. This approach is depicted in the middle and right-hand plots of the image below for the Y resonance.
At the end of this section, then, you will have to make these fits for each bin in the range of interest.
The dataset used in this exercise has been collected by the CMS experiment, in proton-proton collisions at the LHC. It contains 986100 entries (muon pair candidates) with an associated invariant mass. For each candidate, the transverse momentum (pt), rapidity(η) and azimuthal angle (φ) are stored, along with a binary flag probe_isTrkMatch, which is 1 in case the corresponding probe satisfied the track matching selection criteria and 0 in case it doesn’t.
Copy CMSDAS_TP inside CMSSW_14_0_19/src:
cp -r /eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/CMSDAS_TP .
Exploring the content of the TP_Z_DATA.root and TP_Z_MC.root files, the StandAloneEvents tree has these variables in which we are interested in:
Variables to be checked
- pair_mass
- probe_isTrkMatch
- probe_pt
- probe_eta
- probe_phi
We’ll start by calculating the efficiency as a function of the probe η. It is useful to have an idea of the distribution of the quantity we want to study. In order to do this, plot the invariant mass and the probe variables.
Now that you’re acquainted with the data, open the Efficiency.C file. We’ll start by choosing the desired bins for the rapidity. If you’re feeling brave, modify bins for our fit remembering that we need a fair amount of data in each bin (more events mean a better fit!). If not, we’ve left a suggestion in the Efficiency.C file. Start with the eta variable.
Now that the bins are set, we have defined the initial parameters for our fit. In this code, we execute a simultaneous fit using a Voigtian curve for the Z peak. For the background we use a Falling Exponential. The function used, doFit(), is implemented in the source file src/DoFit.cpp and it was based on the RooFit library. You can find generic tutorials for this library here. If you’re starting with RooFit you may also find this one particularly useful. You won’t need to do anything in src/DoFit.cpp but you can check it out if you’re curious.
To get the efficiency plot, we used the TEfficiency class from ROOT. You’ll see that in order to create a TEfficiency object, one of the constructors requires two TH1 objects, i.e., two histograms. One with all the probes and one with the passing probes.
The creation of these TH1 objects is taken care of by the src/make_hist.cpp code. Note that we load all these functions in the src area directly in header of the Efficiency.C code. Now that you understand what the Efficiency.C macro does, run your code with in a batch mode (-b) and with a quit-when-done switch (-q) root -q -b Efficiency.C.
When the execution finishes, you should have 2 new files. One on your working directory Histograms_Data.root and another one Efficiency_Run2018.root located at Efficiency_Result/eta. The second contains the efficiency we calculated, while the first file is used to re-do any unusable fits. If you want, check out the PDF files under the Fit_Result/ directory, which contain the fitting results as the following one:
Now we must re-run the code, but before that, change IsMc value to TRUE. This will generate an efficiency for the MC.
After having both data and MC plot ready, we can make comparison between MC and 2018 run. If so, now uncomment Efficiency.C the following line:
// compare_efficiency(quantity, "Efficiency_Result/eta/Efficiency_Run2018.root", "Efficiency_Result/eta/Efficiency_MC.root");
and run the macro again. You should get something like the following result if you inspect the image at Comparison_Run2018_vs_MC_Efficiency.png:
![]()
If everything went well and you still have time to go, repeat this process for the two other variables, pT and φ! In case you want to change one of the fit results, use the change_bin.cpp function commented in Efficiency.C. If you would like to explore the results having more statistics, use the samples in DATA/Z/ directory!
If you would like to play with the actual CMS tracking workflow have a look at the ntuple generator and the efficiency computation frameworks!
Looking at secondary vertices (continued)
As an exercise, plot the positions of the vertices in a way that corresponds to the CMS half-view. That is, make a histogram like the following:
import math
import ROOT
rho_z_histogram = ROOT.TH2F("rho_z", "rho_z", 100, 0.0, 30.0, 100, 0.0, 10.0)
in which the horizontal axis will represent z, the direction parallel to the beamline, and the vertical axis will represent rho, the distance from the beamline, and fill it with (z, rho) pairs like this rho_z_histogram.Fill(z, rho).
Compute rho and z from the vertex objects:
events.toBegin()
for event in events:
event.getByLabel("SecondaryVerticesFromLooseTracks", "Kshort", secondaryVertices)
for vertex in secondaryVertices.product():
rho_z_histogram.Fill(abs(vertex.vz()), math.sqrt(vertex.vx()**2 + vertex.vy()**2))
rho_z_histogram.Draw()
Question
What does the distribution tell you about CMS vertex reconstruction?
Half-view of CMS tracker (color indicates average number of hits):
![]()
Correlation between pile-up and number of clusters
import ROOT
import DataFormats.FWLite as fwlite
events = fwlite.Events("/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
clusterSummary = fwlite.Handle("ClusterSummary")
h = ROOT.TH2F("h", "h", 100, 0, 20000, 100, 0, 100000)
events.toBegin()
for event in events:
event.getByLabel("clusterSummaryProducer", clusterSummary)
cs = clusterSummary.product()
try:
h.Fill(cs.getNClus(cs.PIXEL),
cs.getNClus(cs.PIXEL) + cs.getNClus(cs.STRIP))
except TypeError:
pass
c = ROOT.TCanvas("c", "c", 800, 800)
h.Draw()
h.Fit("pol1")
c.SaveAs("pileup_nclusters.png")
Harder questions: dxy biases
The dxy parameter is not simply a distance, it is a signed distance. A helical trajectory traces a circle in the plane transverse to the beamline, and the sign of dxy depends on whether the reference point is included inside of that circle our outside of it. You can change the reference point with which dxy is computed by passing a point or a beamspot as an argument:
print( track.dxy())
-0.00082122773835
print( track.dxy(ROOT.math.XYZPoint(0, 0, 0)))
-0.00082122773835
print( track.dxy(beamspot.product()))
-0.00636893586021
Passing no parameter is equivalent to passing (0, 0, 0).
Consider the following script:
import DataFormats.FWLite as fwlite
import ROOT
events = fwlite.Events("/eos/uscms/store/user/cmsdas/2025/short_exercises/trackingvertexing/run321167_ZeroBias_AOD.root")
tracks = fwlite.Handle("std::vector<reco::Track>")
beamspot = fwlite.Handle("reco::BeamSpot")
dxy_vs_phi_000 = ROOT.TProfile("dxy_vs_phi_000", "dxy_vs_phi_000", 100, -3.14, 3.14)
dxy_vs_phi_beamspot = ROOT.TProfile("dxy_vs_phi_beamspot", "dxy_vs_phi_beamspot", 100, -3.14, 3.14)
events.toBegin()
for event in events:
event.getByLabel("generalTracks", tracks)
event.getByLabel("offlineBeamSpot", beamspot)
for track in tracks.product():
dxy_vs_phi_000.Fill(track.phi(), track.dxy(ROOT.math.XYZPoint(0, 0, 0)))
dxy_vs_phi_beamspot.Fill(track.phi(), track.dxy(beamspot.product()))
dxy_vs_phi_000.SetAxisRange(-0.2, 0.2, "Y")
dxy_vs_phi_beamspot.SetAxisRange(-0.2, 0.2, "Y")
c = ROOT.TCanvas("c", "c", 800, 800)
dxy_vs_phi_000.Draw()
dxy_vs_phi_beamspot.SetMarkerColor(ROOT.kRed)
dxy_vs_phi_beamspot.SetLineColor(ROOT.kRed)
dxy_vs_phi_beamspot.Draw("same")
c.SaveAs("beamspot.png")
It produces two plots, one of which shows a clear bias from the correct distribution (on average, dxy should be zero). This bias is not an problem with the detector, only a problem with interpreting it. Can you explain the origin of the bias, including its shape with respect to phi?
Below, there is a figure that might help. It is also useful to draw sketches for yourself.
Bonus Exercise: µµTrkTrk reconstruction in MiniAOD
Please have a look to the instruction at the repository at https://github.com/CMSTrackingPOG/2mu2trk_exercise/blob/main/README.md.
Please notice that it requires a newer CMSSW version!
Documentation
Key Points
At this point it is extremely important to look at the documentation to find out moreabout the Tracking Physics Object Group (TRK POG) tasks.